Abstract
Sacubitril is an ethyl ester prodrug of LBQ657, the active neprilysin (NEP) inhibitor and a component of LCZ696 (sacubitril/valsartan). We report herein the three-dimensional structure of LBQ657 in complex with human NEP at 2 Å resolution. The crystal structure unravels the binding mode of the compound occupying the S1, S1’ and S2’ sub-pockets of the active site, consistent with a competitive inhibition mode. An induced fit conformational change upon binding of the P1’-biphenyl moiety of the inhibitor suggests an explanation for its selectivity against structurally homologous zinc metallopeptidases.
Similar content being viewed by others
Introduction
Sacubitril/valsartan (LCZ696) was recently approved by the European Commission and the U.S. Food and Drug Administration to reduce the risk of cardiovascular death in heart failure patients with reduced ejection fraction1. Nearly 6 million people in the US suffer from heart failure and about half have the reduced ejection fraction form2. LCZ696 (Fig. 1) is a sodium salt complex comprising the anionic forms of the angiotensin II type-1 receptor antagonist valsartan and the NEP inhibitor prodrug sacubitril3. Upon oral administration, LCZ696 delivers rapid systemic exposure to sacubitril and valsartan. Sacubitril is converted by enzymatic cleavage of the ethyl ester to the active metabolite LBQ6574. LBQ657 is a single diastereomer with specific stereocenters (Fig. 1).
Neprilysin, also called neutral endopeptidase, enkephalinase, or atriopeptidase, is a zinc dependent type II integral membrane peptidase belonging to the M13 family. It degrades a variety of vasoactive peptides such as atrial natriuretic peptide, brain natriuretic peptide, bradykinin, adrenomedullin and endothelin-15. NEP inhibition leads to an increased level of these peptides. The combination of NEP and angiotensin receptor inhibition is superior to either agent alone and leads to vasodilation and reduction of extracellular fluid via sodium excretion6.
In recent years, several structures of the soluble extracellular domain of NEP complexed with inhibitors have become available7,8,9,10,11. In this report we describe the crystal structure of NEP in complex with LBQ657 and rationalize the selectivity of the compound relative to the homologous peptidases, endothelin converting enzyme (ECE-1) and neprilysin 2 (NEP2).
Results and Discussion
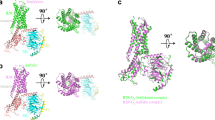
The extracellular domain of human NEP (residues 54–749) was expressed using the baculovirus system. NEP in complex with LBQ657 crystallized in the orthorhombic space group P212121 with two copies in the asymmetric unit diffracting to 2.0 Å resolution. The structure was solved by molecular replacement. The initial difference electron density clearly represents LBQ657 (Fig. 2a). As reported previously7, the extracellular domain of NEP is comprised of two largely α-helical subdomains, with domain 1 predominantly containing C-terminal residues while domain 2 is formed mainly from the N-terminal half of the protein. The two domains are arranged to harbor a large cavity in the center that contains the catalytic machinery which is formed by residues from domain 1 (Fig. 2b). LBQ657 is bound to the active site of NEP by an intricate network of interactions that involves all functional groups of the compound giving rise to the high inhibitory potency of 5 nM12. In the crystal structure, the catalytic zinc atom of NEP is ligated by the side chains of residues His583 (2.0 Å), His587 (1.9 Å) and Glu646 (2.0 Å) with the fourth coordination provided by the carboxylate oxygen adjacent to the P1 methyl of the compound (2.0 Å). The second oxygen atom of this carboxylate is at a distance of 2.7 Å from the catalytic zinc. The methyl group of LBQ657 is pointing towards the shallow S1 pocket making hydrophobic interactions with Phe544. There are two chiral centers in LBQ657 with the specific configuration in both centers facilitating optimal interactions with the active site of NEP. In fact, the other three diastereoisomers of LBQ657 reported by Ksander et al. show a reduction in the potency by about 5 to more than 100-fold12.
Active site view of NEP in complex with LBQ657.
(a) Initial Fo-Fc difference electron density map for LBQ657 is shown in blue mesh. The map is contoured at 2.5 σ. (b) LBQ657 (yellow) makes H-bonding interactions (black dotted lines) with residues of NEP (white carbon atoms) and coordinates with zinc (pink). The active site surface of NEP is shown in grey.
The backbone amide of LBQ657 forms H-bonding interactions with the side chains of Asn542 and Arg717. The amide directionality in LBQ657 is different relative to peptide substrate and inhibitors reported in published crystal structures7,8,9,10,11. In LBQ657, the –NH of the amide is attached to the chiral carbon atom, while in the inhibitors from crystal structures reported previously7,8,9,10,11, the attachment occurs via the amide carbonyl. It is interesting to note that for both types of amide, the nitrogen and oxygen atoms occupy similar positions in the complex structures and are engaged in the same type of interactions (Fig. 3a). The succinic acid in P2’ has been shown to be optimal compared to analogs with shorter or extended chain length12. It places the carboxylic acid in an ideal position for optimal interaction with Arg102 and Arg110, unique for LBQ657.
The P1’ biphenyl of LBQ657 binds deeply in the S1’ subsite of NEP, inducing conformational changes for optimal hydrophobic interactions. A concerted change in conformation of residues Trp693 and Phe106 when a compound containing a biphenyl group is bound in P1’ has been reported previously8. When comparing our co-crystal structure to one with a compound harboring a benzyl in P1’8, an analogous induced fit of Trp693 towards the S2’ subsite is observed, which in turn leads to a conformational change of Phe106 (Fig. 3a). In NEP the space occupied by the new orientation of Trp693 is available because the adjacent Gly714 does not project a sidechain towards this area. LBQ657 does not inhibit angiotensin converting enzyme (ACE), or endopeptidases homologous to NEP such as ECE-1 (39% sequence identity with NEP) and NEP2 (55% sequence identity with NEP) when tested at a maximum concentration of 30 μM (unpublished data). The residues corresponding to Gly714 are Ser735 (ECE-1)13 and Leu744 (NEP2) and their side chains will impose steric constraints to the conformational change of Trp714 (ECE-1) and Trp723 (NEP2), respectively (Fig. 3b). The X-ray structure of ECE-1 confirms Ser735 at the position corresponding to NEP Gly714 (Fig. 3b)10. Additional active site residue differences to NEP will also have an impact on selectivity, such as Arg110NEP →Trp153ECE-1.
All of the molecular interactions between LBQ657 and the enzyme are non-covalent, in line with a reversible inhibition mode. The crystal structure in addition shows the presence of a sub-pocket extending from the meta-position of the second phenyl ring in P1’ to the backbone carbonyl of Arg717.
In conclusion, the reported crystal structure shows how LBQ657 binds to NEP, highlighting the interactions in the catalytic site. The crystal structure unravels the importance of the chiral centers of LBQ657 for optimal binding to NEP, zinc ligation by the carboxylate oxygen adjacent to the P1 methyl and conservation of hydrogen bonding interactions by the backbone amide. An induced fit binding of the bulky biphenyl group in the S1’ subsite sheds light on the plasticity of NEP and yields insights into the mechanism of selectivity. Further work probing the interactions observed in this co-crystal structure, especially the structure activity relationship around the second phenyl ring will be reported separately.
Methods
Protein expression
Human Neprilysin (residues Gly52-Trp749) was expressed using the baculovirus expression system, utilizing a Cathepsin D leader sequence and C-terminal Strep tag. A 10 liter culture of SF21cells was infected at a multiplicity of infection (MOI) of 1.0 and the supernatant harvested after 72 h and frozen at −80 °C.
Protein purification
Four liters of baculovirus expression supernatant were allowed to thaw overnight and any precipitate that had formed was removed by centrifugation at 10,000 g for 15 min. The supernatant, following addition of a 1/10 volume of 10× stock buffer A (10 mM Tris at pH 8, 250 mM NaCl, 1 mM MgCl2 and 1 mM CaCl2), was loaded onto 60 ml column of Con A Sepharose equilibrated with buffer A. After loading the sample, the column was washed with buffer A until the UV absorbance returned to baseline. The column was then eluted directly using a buffer B (buffer A, containing 0.5 M mannopyrannoside and 0.5 M glycopyrannoside). The NEP-containing fractions (450 ml) were dialyzed over the weekend at 4 °C against 25 L of 25 mM Tris at pH 8.0. 350 ml of 3.6 M ammonium sulfate was added to 500 ml of the retentate and following centrifugation (10,000 g; 5 min), the supernatant was loaded onto a 22 ml column of Source™ Phenyl resin which had been equilibrated with buffer C (50 mM Tris, 1.5 M ammonium sulfate). The column was eluted with a gradient of 0–50% buffer D (buffer C without ammonium sulfate) over 30 column volumes with a final step to 100% buffer D. The eluted protein peak was dialyzed against 25 mM Tris at pH 8, containing 1 mM MgCl2 (buffer E) and further diluted with 2 volumes of the same buffer prior to loading onto a 1 ml MonoQ™ anion-exchange column. The column was eluted with a 0–25% gradient of buffer F (buffer E plus 1 M NaCl) over 18 column volumes. The eluted protein peak was pooled with care being taken to exclude small shoulders on the trailing edge of the peak. The pool was concentrated to 15.1 mg/ml using a 5 kDa cut-off centrifugal concentrator.
LBQ657 stock preparation
A stock solution at 100 mM concentration in 100% DMSO was prepared using powder of LBQ657.
Crystallization
For crystallization, the protein was used at a concentration of 10 mg/ml in 20 mM Tris at pH 8.0, 125 mM NaCl, 2 mM MgCl2. LBQ657 (100 mM in DMSO) was mixed with the protein to a final inhibitor concentration of 1mM. Crystallization experiments were performed at room temperature using the hanging drop vapor diffusion method with a 1:1 ratio of inhibitor protein complex mixed with the reservoir solution containing 25% (w/v) PEG3350, 200 mM ammonium acetate, 100 mM BisTris, pH 6.5. Crystals appeared overnight. Before data collection, the crystals were transferred to 25% PEG3350, 200 mM ammonium acetate, 100 mM BisTris at pH 6.5, 20% (v/v) glycerol and flash frozen in liquid nitrogen.
Data collection, structure determination and refinement
X-ray diffraction data were collected at the PXII beamline of the Swiss Light Source (Villigen, Switzerland) at a wavelength of 1.00 Å at 100 K and processed with XDS as implemented in the program package APRV14,15. The crystals diffracted to 2.0 Å resolution and belong to the space group P212121 with two monomers in the asymmetric unit. The structure was solved by molecular replacement using the coordinates of PDB entry 1DMT as a search model7. Iterative cycles of model building and refinement were carried out using programs COOT16, Refmac17 and BUSTER18. The final R-factor is 19.3% (Rfree 22.8%). The data collection and refinement statistics are summarized in Table 1. The coordinates of the refined structure have been deposited in the Protein Data Bank (5jmy).
Additional Information
How to cite this article: Schiering, N. et al. Structure of neprilysin in complex with the active metabolite of sacubitril. Sci. Rep. 6, 27909; doi: 10.1038/srep27909 (2016).
References
McMurray, J. J. V. et al. Angiotensin-Neprilysin inhibition versus Enalapril in heart failure. N Engl J Med 371, 993–1004 (2014).
Mozaffarian, D. et al. Heart disease and stroke statistics-2015 update a report from the American heart association. Circulation 131, E29–E322 (2015).
Feng, L. L. et al. LCZ696: a dual-acting sodium supramolecular complex. Tetrahedron Lett 53, 275–276 (2012).
Gu, J. et al. Pharmacokinetics and pharmacodynamics of LCZ696, a novel dual-acting angiotensin receptor-neprilysin inhibitor (ARNi). J of Clin Pharmcol 50, 401–414 (2010).
Roques, B. P., Noble, F., Dauge, V., FournieZaluski, M. C. & Beaumont, A. Neutral endopeptidase-24.11-structure, inhibition and experimental and clinical-pharmacology. Pharmacol Rev 45, 87–146 (1993).
McMurray, J. J. V. Neprilysin inhibition to treat heart failure: a tale of science, serendipity and second chances. Eur J Heart Fail 17, 242–247 (2015).
Oefner, C., D’Arcy, A., Hennig, M., Winkler, F. K. & Dale, G. E. Structure of human neutral endopeptidase (neprilysin) complexed with phosphoramidon. J Mol Biol 296, 341–349 (2000).
Oefner, C., Roques, B. P., Fournie-Zaluski, M. C. & Dale, G. E. Structural analysis of neprilysin with various specific and potent inhibitors. Acta Crystallogr Sect D-Biol Crystallogr 60, 392–396 (2004).
Sahli, S. et al. Second-generation inhibitors for the metalloprotease neprilysin based on bicyclic heteroaromatic scaffolds: synthesis, biological activity and X-ray crystal-structure analysis. Helv Chim Acta 88, 731–750 (2005).
Oefner, C., Pierau, S., Schulz, H. & Dale, G. E. Structural studies of a bifunctional inhibitor of neprilysin and DPP-IV. Acta Crystallogr Sect D-Biol Crystallogr 63, 975–981 (2007).
Glossop, M. S. et al. Synthesis and evaluation of heteroarylalanine diacids as potent and selective neutral endopeptidase inhibitors. Bioorg Med Chem Lett 21, 3404–3406 (2011).
Ksander, G. M. et al. Dicarboxylic-acid dipeptide neutral endopeptidase inhibitors. J Med Chem 38, 1689–1700 (1995).
Schulz, H., Dale, G. E., Karimi-Nejad, Y. & Oefner, C. Structure of human endothelin-converting enzyme I complexed with phosphoramidon. J Mol Biol 385, 178–187 (2009).
Kroemer, M., Dreyer, M. K. & Wendt, K. U. APRV-a program for automated data processing, refinement and visualization. Acta Crystallogr Sect D-Biol Crystallogr 60, 1679–1682 (2004).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26, 795–800 (1993).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D-Biol Crystallogr 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D-Biol Crystallogr 53, 240–255 (1997).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr Sect-D Biol Crystallogr 68, 368–380 (2012).
Acknowledgements
The authors would like to dedicate this paper to the memory of Gary Ksander.
Author information
Authors and Affiliations
Contributions
P.R. and C.L. produced and purified the proteins. A.D. and F.V. performed crystallization experiments. N.S. collected crystallization data and solved the structure. F.C. determined inhibitory activity of the compounds, G.M.K. designed the compounds, conceived the experiments, supervised the work. R.G.K., N.S., C.W. and M.M. interpreted data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Schiering, N., D’Arcy, A., Villard, F. et al. Structure of neprilysin in complex with the active metabolite of sacubitril. Sci Rep 6, 27909 (2016). https://doi.org/10.1038/srep27909
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27909
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.