Abstract
Lipocalins are one of the most important groups of inhalant animal allergens. The analysis of structural features of these proteins is important to get insights into their allergenicity. We have determined two different dimeric crystal structures for bovine dander lipocalin Bos d 2, which was earlier described as a monomeric allergen. The crystal structure analysis of all other determined lipocalin allergens also revealed oligomeric structures which broadly utilize inherent structural features of the β-sheet in dimer formation. According to the moderate size of monomer-monomer interfaces, most of these dimers would be transient in solution. Native mass spectrometry was employed to characterize quantitatively transient dimerization of two lipocalin allergens, Bos d 2 and Bos d 5, in solution.
Similar content being viewed by others
Introduction
A characteristic feature for a type I allergy is the development of a special class of IgE antibodies against a group of proteins which are called allergens1. It has been observed that known allergens are members of a limited class of protein families, which suggests that structural features of these proteins would contribute to allergenicity. It is also known that the immune system develops IgE antibodies in parasitic worm (helminths) infections. Recently, structural resemblances between allergens and helminth antigens2 have been pointed out. In immediate phase reaction allergens, cross-link allergen-specific IgE antibodies bind to the FcεRI receptors on the surface of mast cells or basophils. This leads to the degranulation and release of pharmacologically active mediators, such as vasoactive amines.
The cross-linking requires that the allergen is at least bivalent, displaying two conformational B-cell epitopes to which IgE antibodies would bind. If the allergen is monomeric and thus asymmetric, this would require two different IgE antibodies and, consequently, two different epitopes for the allergen. However, if the allergen is oligomeric this would allow a multifold presentation of a monovalent epitope and, consequently, requires only one allergen-specific IgE antibody. We have previously characterized some common allergens and suggested that oligomerization is a common feature among allergens. Many allergens are transient dimers in solution, whose monomer-dimer ratio is dependent on protein concentration and the dissociation constant (KD)3. In order to investigate the prevalence of dimerization, we have studied lipocalins, one of the major allergen families.
Lipocalins include the most important inhalant animal allergens. In addition, a bovine milk allergen (Bos d 5) and several arthropodan allergens are lipocalins. Lipocalins are typically small proteins (about 200 amino acid residues) composed of an 8-stranded up-and-down antiparallel β-barrel and one α-helix. There is a hydrophobic cavity inside the β-barrel capable of binding a hydrophobic or amphiphilic ligand (Fig. 1a). In most cases, the native ligands for lipocalins are unknown4,5. In order to analyze the oligomeric structures of lipocalins, we have studied all 10 crystal structures of lipocalin allergens in the Protein Data Bank (PDB). Bovine dander allergen Bos d 2 was the only one found to be monomeric6. Because the protein had been crystallised at an acidic pH (3.4–4.5)7, we decided to search for a new crystal form at a more neutral pH. In fact, both new crystal forms of Bos d 2 contained a symmetric weak dimer. The existence of a dimeric Bos d 2 in solution was observed by native mass spectrometry (MS). Furthermore, we used native MS to quantify the dimerization for Bos d 2 and Bos d 5. In the following, we present an analysis of oligomeric structures of lipocalin allergens and discuss their implications for the presentation of multiple epitopes and the triggering of FcεRI bound IgE antibodies on mast-cells and basophils.
(a) The ribbon representation for monomeric Bos d 2. The eight antiparallel β-strands (A–H) forming a central barrel are shown as cyan arrows. The β-strand (I) outside the barrel is in green. The α-helix is in red and loops connecting different secondary structure elements are labelled L1 to L8. The black arrow shows the entrance to the ligand binding pocket (b) The monomers of 9 different lipocalin allergens which form symmetric homodimers are superimposed to observe the orientation of the second monomer. The superimposed monomers are in green and grey, blue and red colours are used for the second monomers.
Results
Dimeric crystal structures of Bos d 2
The search for new crystal forms for bovine allergen Bos d 2 at neutral pH resulted in two new crystal forms. Monoclinic crystals diffracted at a 1.4 Å resolution. The asymmetric unit contained one Bos d 2 molecule but the space group C2 includes a 2-fold symmetry axis, consequently, half of the dimer can be formed by a crystallographic symmetry operator. Trigonal crystals diffracted at 1.75 Å resolution. Again, the asymmetric unit contained one Bos d 2 molecule and a dimer can be formed with the use of a crystallographic symmetry operator. Bos d 2 contains a central β-barrel composing of eight antiparallel β-strands (A-H). In addition, there is a short β-strand (I) as well as an α-helix (Fig. 1a). The r.m.s. differences of Bos d 2 molecules in orthorhombic and monoclinic crystals is 0.226 Å, between orthorhombic and triclinic crystals there is 0.297 Å and between monoclinic and triclinic crystals there is 0.242 Å, which are in the range observed for different molecules in the asymmetric unit in protein crystal structures. The largest changes in the structures are located in the L3 loop and the L8 loop that connects the C-terminus of β-strand H to the α-helix (Fig. 2e).
Dimerization of Bos d 2 observed in crystals as a ribbon presentation.
(a) The symmetric dimer of Bos d 2 observed in the monoclinic (C2) crystal form, (b) in the trigonal (P3221) crystal form. The key residues which contribute to the binding on the monomer-monomer interface in Bos d 2 dimer in monoclinic (c) and in trigonal (d) crystal forms. (e) The superimposition of a Cα-backbone of monomeric Bos d 2 (orthorhombic, in red), dimeric Bos d 2 (monoclinic, in cyan) and dimeric Bos d 2 (trigonal, in grey).
The monomer-monomer interface area for the dimer of Bos d 2 in a monoclinic crystal is 389 Å2. The interface consists of a terminus of β-strands E-H and loops L5 and L7 (Fig. 2a). Amino acid residues Thr85 and Tyr105 along the 2-fold axis pack against the same residues from the second monomer (Fig. 2c). Glu81, located on the interface, has multiple conformations. In addition, Gln73 makes a hydrogen bond with the main-chain carbonyl of Gly107 and the side-chain of Val79 packs against Gly107. The interface area between monomers in the trigonal crystal is also 389 Å2 but there are many more hydrophobic contacts on the monomer-monomer interface. The side-chains of Thr62, Leu64 and Leu66 from the β-strand D form the core of the interface. In addition, the residues Phe82 and Tyr83 from the L5 loop, as well as Leu57 from the β-strand C, are located on the interface (Fig. 2b,d). Because there is a clearly higher number of hydrophobic residues on this interface, we assume that this interface is more favourable in solution than the dimer observed in monoclinic crystal form.
Analysis of crystal structures of lipocalin allergens
In addition to Bos d 2, there are three-dimensional coordinates for nine other allergens in the PDB. Seven allergens are from mammals and three from arthropods (Table 1).
Rat n 1 allergen is a major urinary protein of male rats. The tetrameric structure of Rat n 1 has been reported in monoclinic and orthorhombic crystal forms8,9. The structure contains a complete tetramer with 222 symmetry, each monomer packs against two other monomers having two different interfaces. The larger monomer-monomer interface (708 Å2) is formed by strand D-F and especially by the L4 loop between β-strands D and E (Fig. 3a). Three aromatic hydrophobic residues Tyr72, Phe81 and Phe108 can be found on the interface, which suggests strong hydrophobic interactions, which would favour oligomer formation. The second interface is smaller (457 Å2) and formed mainly by a β-strand C. Tyr86 from this β-strand packs with the same residue from the second monomer. Because the second interface is smaller with fewer hydrophobic interactions, this would indicate that tetramer dissociates easily to dimers at lower protein concentrations.
Can f 2 allergen has been reported to be a dimeric protein10 but, when describing the crystal structure, Madhurantakam et al. consider that Can f 2 is monomeric in neutral pH11. A symmetric dimer has been described with a 447 Å2 interface area12. However, the same crystal structure also shows a tetrameric arrangement in which one interface (576 Å2) is formed between β-strands C and D from one monomer and β-strand I and the α-helix from the second monomer (Fig. 3b). Phe72 on the outer surface of the β-strand D packs with the hydrophobic Leu131 and Val50 from the second monomer. The second interface is smaller (380 Å2) and it is formed between N-terminal residues and the C-terminus of β-strand F from one monomer and loops L1 and L3 from the second monomer. Trp35 from loop L1 interacts with Val95 and Pro97 from another monomer. There is no symmetry axis between these interfaces but there is one 2-fold axis through the whole tetramer (Fig. 3b).
Can f 4 is the second dog lipocalin allergen, whose crystal structure has been determined at 2.6 Å resolution. The asymmetric unit contained one monomer and one symmetric dimer in which the monomer-monomer interface area is 640 Å2. Hydrophobic residues located on β-strands E-H form the core of the dimeric interface (Fig. 3c). These include Leu76, Tyr86, Val102, Val104 and Ile11012.
Mus m 1 is a mouse major urinary protein, whose structure was determined at 2.4 Å resolution and on the basis of the crystal structure it was deduced to be a dimer8. The monomer-monomer interface area is smaller than other lipocalins allergens (389 Å2) but it contains a cluster of hydrophobic residues, Leu 52, Val57 and Val74, as well as a salt-bridge (Lys59-Asp76), which are all located in β-strands F and G (Fig. 3d).
The crystal structure for Equ c 1 was determined at 2.3 Å resolution with one molecule in the asymmetric unit13. The monomer-monomer interface area is the largest among the lipocalin allergens (1025 Å2). As in Can f 4, the interface is formed by β-strands F, G and H but loop L6 has a larger role on the interface (Fig. 3e). The hydrophobic residues Val93, Phe112, Y108 and V110 are located on this interface.
Bos d 5, or β-lactoglobulin, is a major protein in bovine milk (concentration ca. 3–4 mg/ml). On the basis of its crystal structure, Bos d 5 has regularly been described as a dimer, although the interface area is only 528 Å2. The structure has been solved at 1.8 Å resolution with one dimer in the asymmetric unit14. Dimerization is achieved by main-chain hydrogen bonds between β-strands I and participation of some N-terminal residues (Fig. 3f). Hydrophobic residues Ile29, Ile147, Leu139 and Phe151 can be found on the interface. Bos d 5 also existed as a dimer in complex with Fab fragments of an IgE antibody15.
Per a 4 is a lipocalin allergen from the American cockroach (Periplaneta americana). On the basis of gel-filtration, it was deduced that Per a 4 would exist as a dimer in solution. The proposed dimer consisted of both molecules in the asymmetric unit (interface 663 Å2), having some hydrophobic contacts, but the dimer is asymmetric16. However, the crystal structure also shows a two-fold symmetry axis between the two molecules in the asymmetric unit. The interface area for this dimer is smaller (403 Å2), but there is a cluster of hydrophobic Ile65 and Ile69 from the β-strand D and loop L3. In addition, there are main-chain hydrogen bonds between β-strands D (Fig. 3h).
Arg r 1 is an allergen from the European pigeon tick, whose bite can cause anaphylactic reactions in sensitized humans17. The crystal structure of Arg r 1 has been determined at high resolution (1.4 Å) but there is no structure paper. The crystal structure contains three molecules in the asymmetric unit in which two molecules form a dimer and the third one forms a similar dimer with a crystallographically symmetric molecule. The monomer-monomer interface is 530 Å2. The residues from the N-terminal and the L8-loop between the β-strand H and the α-helix are located on the interface (Fig. 3i). There are four hydrogen bonds between the main-chains and other polar contacts but not very many hydrophobic contacts. However, the existence of two similar dimers in crystal suggests that this dimer is quite favourable.
Bla g 4 is also a cockroach allergen but from a different species (Blattella germanica). Bla g 4 is assumed to be a monomer in solution, according to gel-filtration results16. However, two molecules in the asymmetric unit form a dimeric structure having a quite large monomer-monomer interface (842 Å2), which is formed by a L8-loop between the β-strand H and the α-helix and L3-loop (Fig. 3j). A large number of hydrophobic residues, Ile46, Met 127, Leu 129, Phe 132, Met141 and Ile142, on the interface support the dimer formation.
Comparison of lipocalin allergen oligomeric structures
All the oligomeric structures of lipocalin allergens are different to each other. This is probably due to the fact that amino acid identity between them is generally low (5–20%). The highest identity is between Mus m 1 and Rat n 1 allergens (64%) but they still have different oligomeric structures. Different oligomerization can be explained by variation of amino acid residues on the protein surfaces. Dayhoff et al. have observed that oligomers would have the same binding mode if sequence identity would be 70% or higher between them18. In order to see different modes for dimerization, we have superimposed the symmetric dimeric structures of allergens in a way that the position for one monomer is fixed (Fig. 1b). Although the position of the second monomer varies considerably, there are no monomers bound at both ends of the central β-barrel. Consequently, the oligomerization would not prevent ligand binding or release. Dimerization of Bos d 2 and Per a 4 resemble each other: both lipocalins form an extended β-sheet through β-strand D, although there are also some differences in the packing of these β-strands. In addition, the dimers of Can f 4 and Equ c 1 are quite similar: they both have a similar packing of β-sheets together15. We can also notice that the majority of the lipocalin allergens utilize inherent features of a β-sheet in the formation of symmetric dimers. Bos d 5, Bos d 2 and Per a 4 form an extended β-sheet by joining the β-sheets from two monomers together. In Bos d 2 and Per a 4, this is mediated by β-strand D, in Bos d 5 by β-strand I. On the other hand, in dimerization, Can f 2, Can f 4, Equ c 1, Mus m 1, and Rat a 1 form a new β-sandwich structure by bringing β-sheets from two monomers together. In addition, the monoclinic structure of Bos d 2 utilizes similar β-sheet packing. The exceptions are oligomeric structures of Bla g 4 and Arg r 1, which are predominantly joined together by long L8 loops. The observation that β-sheets have an essential role in the formation of dimers suggests that the inherent features of β-sheets (packing of β-strands and extension of β-sheets) also have a role in the formation of weak dimers and that there is a limited number of arrangements to form transient dimers. In consequence, the packing features at β-sheets can perhaps also be used to analyze different protein-protein interfaces in crystals in order to distinguish native interactions and crystal contacts.
Native mass spectrometry of lipocalin allergens
The accurate molecular mass of recombinant Bos d 2 (rBos d 2) was determined by high-resolution Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry in denaturing solution conditions (data not presented). The experimentally determined molecular mass was 17845.89 ± 0.03 Da, which agrees perfectly with the theoretical mass calculated from the amino acid sequence of a mature Bos d 2 with two disulfide bonds (17845.81 Da). Apart from the main proteoform, the mass spectrum revealed that a substantial amount of the protein molecules contained a pyrrolidone carboxylic acid (PCA), formed through a cyclization of the N-terminal glutamine residue (17828.90 Da). Repeated measurements showed that the PCA-containing proteoform started to dominate during long-term storage. The sequence of rBos d 2 contains two methionine residues. The analysis also revealed that the protein was partly oxidized, probably through a single methionine residue and thus displayed a +16 Da mass difference when compared to the main proteoform (17861.90 Da).
Native MS was used to study the dimer formation of rBos d 2 in native-like solution conditions (i.e., 10 mM ammonium acetate, pH 6.9). The basic protocol for native MS was similar to our previous work3, which allows for the weak transient protein dimers to be detected. The native MS spectrum of rBos d 2 (10 μM in respect to the protein monomer) at pH 7 showed signals of both monomers and dimers in solution (Fig. 4a). When the protein concentration was increased to 90 μM, the relative intensity of the dimer increased accordingly (Fig. 4b). These results indicate that rBos d 2 is indeed a weak transient dimer in solution. To further quantify the dimerization (see Methods for details), we obtained protein monomer–dimer ratios from the native mass spectra over a wide range of protein (monomer) concentrations (0–90 μM). The fitted curve of the free monomer concentration against the total protein concentration is presented in Fig. 5a. The determined value for the equilibrium dissociation constant (KD) for self-association was KD = 340 ± 20 μM. We also analyzed the pH-dependence of the dimerization (Fig. 5c) and the amount of rBos d 2 dimer never exceeded the amount of the monomer. The amount of dimer slowly decreased when the pH changed to be more alkaline. The maximal amount of dimer was present at around pH 3.
Native mass spectra of lipocalin allergens at different protein concentrations.
(a,b) rBos d 2 at protein (monomer) concentrations of 10 and 90 μM. The peaks representing the protein monomer (M) are in blue, while the peaks representing the protein dimer (D) are in red. The most abundant peaks for the monomer and the dimer have been assigned with the number indicating the ion charge state (i.e., M7+ = [rBos d 2 + 7H]7+). (c-h) Native mass spectra of Bos d 5 at a protein (monomer) concentration of 1-25 μM.
Protein monomer concentration as a function of total protein concentration for (a) rBos d 2 and (b) Bos d 5 as calculated from the native mass spectra.
The solid lines are the best fits of the data to Equation 4 (see, Online methods for details) for determining KD value for dimerization. Relative abundances of the protein monomer and the dimer for (c) rBos d 2 and (d) Bos d 5 depending on solution pH at a protein (monomer) concentration of 40 μM.
For comparison and validation, we analyzed bovine milk β-lactoglobulin (Bos d 5), another dimeric lipocalin allergen, by native MS in a range of protein monomer concentrations (Fig. 4c–h). The determined equilibrium constant for Bos d 5 self-association was KD = 9.9 ± 0.6 μM (Fig. 5b). Thus, Bos d 2 is clearly a weaker transient dimer than Bos d 5. It must be noted that the KD determined for Bos d 5 represents an apparent value, as there is a mixture of three dimers in solution (AA, AB and BB), due to the presence of two natural variants, A and B. We did not attempt to measure individual KDs for AA, AB and BB, since this would require solving a complex set of equilibrium equations and the native MS data may not be sufficient to derive accurate values for all three KD values. Moreover, the apparent KD reflects better the situation for natural Bos d 5, which is in dynamic equilibrium between A, B, AA, AB and BB. However, relative intensities suggest that AA forms a somewhat weaker transient dimer (i.e., higher KD) than BB. Furthermore, the pH profiles for the dimerization between rBos d 2 and Bos d 5 were slightly different. The amount of Bos d 5 dimer was highest at pH 4–5 (>90%) and it continuously decreased from pH 5 to 8 (Fig. 5d). Above pH 8, the amount of dimer decreased rapidly, due to protein denaturation. Also, the amount of dimer rapidly decreased at pH < 4. The results are in line with the results of Invernizzi et al.19.
Because of its easy availability, biophysical properties of Bos d 5 have been studied extensively. Dimerization of Bos d 5 has been studied by several different analytical methods, including isothermal titration calorimetry (ITC), analytical ultracentrifugation (UAC) and native MS19,20,21,22,23,24. It has been shown that Bos d 5 is a transient dimer in a wide range of solution conditions. For example, the Kd values for the two variants A and B, determined by dilution-ITC at 20 °C and pH 7, are 24 and 14 μM, respectively. This is fully consistent with our present data, as well as our earlier work3. However, dimerization of Bos d 5 is highly dependent on pH, ionic strength and temperature and a range of different Kd values (∼1–200 μM) have been reported for both the variants in different conditions. Most studies have dealt with either of the natural variants of Bos d 5, A or B. However, natural Bos d 5 exists as a mixture of these two variants and the predominant dimer in solution is a heterodimer, AB3. The apparent KD of 9.9 μM for Bos d 5, determined in this work is slightly lower than the values determined earlier for both variants by using dilution-ITC20,21, but clearly higher than determined for the A variant (KD = 2.2 ± 0.7 μM) by Liu et al. using native MS23. There is also evidence that higher oligomeric states (e.g., octamers) can form around a pH close to the isoelectric point of the protein (∼4.6) and at low temperature25. Based on the dilution-ITC data, the dimerization of Bos d 5 is both enthalpy- and entropy-driven, although the enthalpy contribution is more dominant for the association free energy in the variant A22. This is consistent with the monomer-monomer interface formed through both hydrogen bonds, as well as with hydrophobic interactions.
We have earlier introduced the use of native mass spectrometry into studying dimerization of proteins in solution3,15 and develop it further in this study. The obtained results for Bos d 5 are in good agreement with the literature. The results suggest that native MS is able to detect and even quantify transient dimerization of proteins in solutions having KD values in the high-μM to low-mM range.
Discussion
Proteins which exist in a monomer-dimer equilibrium in solution can be defined as weak or transient dimers. Both the Kd value for the dimer and the protein concentration affect the degree of oligomerization (i.e., whether the protein is preferentially in the dimeric or the monomeric form). In addition, ionic strength and pH can play significant roles as well and temperature markedly affects those interactions that are mainly entropy-driven. Typically, Kds are in the micromolar range for transient dimers when the monomer-monomer interface is less than 1000 Å2 26. Transient protein oligomerization is difficult to study experimentally. For example, size-exclusion chromatography (SEC) may suffer from protein interactions with the column matrix, which results in an erroneous estimation of molecular weight and can also shift the equilibrium towards a monomeric form27. Dynamic/static light scattering (DLS/SLS) or low angle X-ray scattering (SAXS) have not been widely used in the study of transient dimers, probably due to the challenges in measuring mixtures of different sizes of particles in solution. ITC and AUC are better suited for the purpose and ITC also directly provides estimates of the association enthalpy and entropy. However, ITC suffers from rather low sensitivity and requires high intial concentrations (for dilution-ITC experiments). Furthermore, AUC requires a priori knowledge of the oligomeric state for the model used to fit the data. Native MS has several benefits over the other techniques; it provides direct stoichiometric data on different oligomeric forms present simultaneously in solution, is highly sensitive and also provides quantitative data for the estimation of Kd values. It is also not inherently affected by association/dissociation kinetics, like many other techniques, since it measures a time-averaged thermodynamic equilibrium. Recently, native MS has been utilized in the determination of Kd values for Bos d 2 and Bos d 5 (340 and 9.9 μM; this study) and Can f 4 (27 μM)15 allergen dimers. All these values of KD are in the micromolar range, typical for transient dimers. The size of the monomer-monomer interfaces varies between 389 Å2 to 1025 Å2 in the 10 studied lipocalin allergen crystal structures (Table 1) suggesting all these dimers could be transient26. It also seems that Equ c 1, which has the largest monomer-monomer interface, is a transient dimer because it was suggested that Equ c 1 exists as a mixture of monomers and dimers in solution28.
The capability of allergens to form dimers or higher oligomers could have a remarkable effect on mast-cell triggering, because a monovalent allergen could present a single epitope two or more times and only a single allergen specific IgE antibody is needed. In theory, if an allergen forms a dimer, only one monoclonal IgE antibody is needed for mast-cell triggering. In consequence, allergen oligomerization could decrease the need for polyclonal IgE antibodies, which, in turn, would have implications on the nature of the epitope repertoire of the IgE response29.
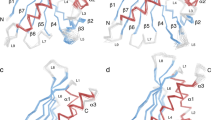
If we sketch the initial process leading to the mast-cell degranulation we can first note that serum IgE antibodies bind very strongly to the FcεRI receptors on the surface of mast cells or basophils (KD ∼ 0.1 nM) (Fig. 6a). In addition, the binding of allergens to IgE antibodies is also potent (KD ∼ 0.1–1.0 nM) (Fig. 6b), generally more potent than antigen binding to IgG antibodies1,15. Both these interactions are many order of magnitudes stronger than transient dimerization of allergens (KD ∼ 10 μM) in solution. Therefore, we can assume that IgE antibodies are predominantly bound to the FcεRI receptors on the cell surface and the allergen prefers to bind as a monomer to the surface bound specific high-affinity IgE-antibodies (Fig. 6b). We could consider that this process leads to colocalization30 of allergens, thus they increase their “concentration” on the surface of mast cells, allowing the dimerization to occur (Fig. 6c,d).
The sketch for the initial events leading to the allergen triggered signal transduction.
(a) allergen-specific IgE serum antibodies bind to FcεRI receptors on the surface of mast cells or basophils with high affinity. (b) allergen exposure leads to a binding of monomeric allergens to specific IgE antibodies already bound to FcεRI receptors. (c) tethering of monomeric allergens on the cell surface results in dimerization of allergen monomers. The 2-dimensional dissociation constant for allergen dimers on the cell surface is not known. (d) the allergen dimerization has cross-linked FcεRI bound IgE antibodies that lead to the signal transduction.
In fact, allergen dimerization on surfaces of mast cells has a clear resemblance to many other signal transduction processes. In a recent study, dimerization of a lipid-anchored H-Ras protein on the membrane surfaces was studied with a combination of biophysical methods and the KD(2D) for the dimer was found to be about 1000 molecules/μm2. The authors estimated that this would correspond to KD being ∼500 μM in solution31, which indicates a clearly weaker dimer than lipocalin dimers. It is clear that in order to prove the role of allergen dimerization for the clustering of FcεRI bound IgE antibodies on the surface of mast cells or basophils substantial further experimentation on the membrane surfaces is required. This study, which is based on the analysis of three-dimensional structures of allergens and their cabability to form dimers in solution, should be considered as the first step in continuing efforts to understand the molecular basis of allergenicity and how structural features of allergens may be related to the signal transduction processes.
Methods
Mass spectrometry of Bos d 2 and Bos d 5
Recombinant Bos d 2 was expressed and purified as reported earlier7. Native Bos d 5 from bovine milk was obtained as a lyophilized powder from Sigma-Aldrich (St. Louis, MO, USA). All mass spectrometric analyses were carried out by a 12 T APEX-Qe FT-ICR mass spectrometer (Bruker Daltonics, Billerica, MA, USA), equipped with an ESI source (Apollo-II). The analysis in denaturing solution conditions was performed with a rBos d 2 sample diluted to 2 μM with MeCN:H2O:HOAc (49.5:49.5:1.0, v/v) and the solution was injected into the ion source by a syringe pump with a flow rate of 1.5 μl/min. Nitrogen was used as the drying (250 °C, 6 mbar) and nebulizing gas. Ions from an ESI-source were accumulated in the hexapole ion trap for 1.0 s and transferred to the ICR cell for trapping, excitation and detection. For a mass spectrum, a total of 256 co-added (1 Mword) time-domain transients were recorded and subjected to fast Fourier transform and magnitude calculation. Mass spectra were externally calibrated by using the ions of an ES Tuning Mix (Agilent Technologies, Santa Clara, CA, USA). For native MS experiments, protein samples were desalted and exchanged into a 10 mM ammonium acetate buffer (pH 6.9) with the PD-10 columns (GE Healthcare Bio-Sciences, Uppsala, Sweden). The instrumental parameters were carefully optimized to maintain weak non-covalent interactions in the gas-phase and to avoid any unintentional oligomer dissociation upon ionization/desolvation3. A data set contained 300 co-added, 256 kword time-domain transients. The dimerization of Bos d 2 and Bos d 5 were studied in the pH range 3–9. Proteins were measured at a (protein monomer) concentration of 1-90 μM in a 10 mM ammonium acetate buffer, whose pH was adjusted either by ammonium hydroxide or acetic acid. In order to avoid changes in the desired pH, caused by the adding of the sample buffer to the pH-adjusted buffer, highly concentrated protein solutions were used (454 μM for BLG and 323 μM for Bos d 2). Moreover, the possible pH changes were further tested on a larger scale by using the same dilution ratio of the sample buffer and the pH-adjusted buffer and by measuring the final pH of the combined solutions. To ensure a valid pH for each measurement, native mass spectra were first recorded from pH 7.0 to pH 3.0 and then from pH 7.0 to pH 9.0. Between the measurements, the sample transfer capillary was carefully flushed with the following pH buffer. Two or three parallel mass spectra were measured for each different pH value. The monomer-dimer ratio was obtained from the absolute intensities of the ions representing either the monomer (P) or the dimer (P2), by combining the intensities (i) of the ions appearing in different charge states. Assuming the same ionization and transmission efficiencies for the monomer and the dimer23, their intensity ratio sufficiently reflects the concentration ratio in solution. The same protocol was used when the dimerization of Bos d 2 was studied as a function of the ionic strength. To determine KD for the dimerization of both proteins, the protein concentration was varied from 0 to 90 μM and the monomer-dimer ratio was obtained. The KD value for protein dimerization can be defined as

where [P] and [P2] are the equilibrium concentrations of the free protein monomer P and protein dimer P2, respectively. We can define the total protein (monomer) concentration [P]total as

Inserting the above expression to the equation (1), we find that

Finding roots for this second-order polynomial gives the expression

which defines [P] as a function of [P]total, if Kd is known. By fitting [P] as a function of [P]total, one can determine Kd in a wide range [P]total by a nonlinear least-squares fitting procedure. A native mass spectrum for a transient dimer directly provides the intensity ratio of P and P2, which can be used to calculate [P] as

Total intensities for P and P2 (iP and iP2) can be calculated as sums of peak intensities iz over the charge state distribution (z = 1 to n) in the ESI-MS spectra. Fitting [P] against [P]total using equation (4) gives KD. Equation (5) is valid only if ionization and ion transmission efficiencies are considered the same for P and P2. In the present and in our previous work3 we optimized all ion source and ion optics parameters to obtain similar transmission efficiency for both the monomer and the dimer. We used Bos d 5 as a reference protein for which KD is known. Liu et al. have previously shown that ionization efficiencies for the monomer and the dimer of Bos d 5 are roughly the same23, hence the use of equation (5) is therefore warranted.
Crystallisation of Bos d 2
The rBos d 2 allergen was previously crystallised in the space group P212121 by using 10% (w/w) PEG6000, 0.1 M sodium acetate (pH 3.4–4.5) as a precipitant solution. The protein solution was concentrated up to 50 mg/ml for crystallisation experiments7. Alternative conditions for crystallisation of rBos d 2 were searched for by using several commercially available crystallisation screens. Protein crystals belonging to space group P3221 were obtained by using a 4.0 M sodium formate (pH 7.4) as a precipitant solution. Single crystals were obtained after 2 months of preparation of crystallisation drops. Protein was crystallised in the space group C2 by using the precipitant solution containing 2.5 M 1,6-hexanediol and 0.1 M sodium citrate (pH 6.4). The growing time for monoclinic crystal was 6 months. Both new crystal forms were obtained with a hanging-drop vapour-diffusion method at 293 K. Crystallisation drops were prepared by mixing 2 μl of protein solution (9 mg/ml in 50 mM ammonium acetate buffer, pH 6.7) and 2 μl of precipitant solution.
For the data collection, P3221 crystals were cryoprotected with a 3.6 M sodium formate, 10% glycerol solution before freezing them in liquid nitrogen. The high hexanediol concentration of the precipitant solution enabled the direct transfer of C2 crystals from the crystallisation drops to liquid nitrogen. Both diffraction data sets were collected by using synchrotron radiation at the ESRF, Grenoble, France. The P3221 crystal diffracted to a resolution of 1.75 Å on beamline ID29. For a complete data set, 900 diffraction images with a 0.1° oscillation range were collected with a Pilatus 6M pixel detector (Dectris Ltd). The diffraction data from the C2 crystal were collected on beamline ID14-4. The crystal diffracted to a resolution of 1.4 Å and the reflections were recorded with a Q315r ADSC detector. The crystal was rotated 180° with an oscillation angle of 0.5°. The data sets were processed with a XDS program and scaled and merged with XSCALE32.
Structure determination and refinement
The packing of protein molecules in the new crystal forms was studied by using the structure of rBos d 2 (PDB code 1BJ7) as a search model. The molecular replacement, carried out with the program Phaser33 as a part of the CCP4 suite34, indicated that both P3221 and C2 crystal forms contained one rBos d 2 molecule in an asymmetric unit. The solvent content of the crystals was 72% and 48%, respectively. The protein models were built manually with the program Coot35 and refined with the program PHENIX36. The protein model in the space group P3221 was refined to final Rwork and Rfree values of 21.1% and 23.2%, respectively. The corresponding values for the rBos d 2 model in the space group C2 were 19.6% and 21.4%. The final structures were validated with the program Procheck. The data collection and refinement statistics are presented in Table 2. Protein structures have been deposited in the RCSB Protein Data Bank with the accession codes of 4wfu and 4wfv.
Analysis of crystal structures
The coordinates for the lipocalin allergens were from the Protein Data Bank (www.rcsb.org). PDBePISA (Proteins, Interfaces, Structures and Assemblies) (http://www.ebi.ac.uk/pdbe/pisa) was used to analyze protein-protein interfaces in crystals37. However, it should be noted that the program is not suitable for estimating the existence of transient dimers because it does not use protein concentration as a parameter. However, PISA is useful in analyzing protein-protein interfaces. In the estimation of the existence of a quaternary structure we used the area of monomer-monomer interface, the symmetry axis between monomers and the nature of interactions (hydrogen bonds, hydrophobic contacts) between monomers. The figures of the proteins were created with the PYMOL program [Delano, W. L. The PyMol Molecular Graphics System, www.pymol.org].
Additional Information
Accession codes: The atomic coordinates and structure factors (codes 4WFV and 4WFU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
How to cite this article: Niemi, M. H. et al. Dimerization of lipocalin allergens. Sci. Rep. 5, 13841; doi: 10.1038/srep13841 (2015).
References
Gould, H. J. & Sutton, B. J. IgE in allergy and asthma today. Nature Rev. Immunol. 8, 205–217 (2008).
Fitzsimmons, C. M., Falcone, F. H. & Dunne, D. W. Helminth allergens, parasite-specific IgE and its protective role in human immunity. Frontiers Immunol. 5, 61 (2014).
Rouvinen, J. et al. Transient dimers of allergens. PLoS ONE 5, e9037 (2010).
Virtanen, T., Kinnunen, T. & Rytkönen-Nissinen, M. Mammalian lipocalin allergens—insights into their enigmatic allergenicity. Clin. Exp. Allergy 42, 494–504 (2012).
Hilger, C. et al. IgE-mediated anaphylaxis caused by bites of the pigeon tick Argas reflexus: Cloning and expression of the major allergen Arg r 1. J. Allergy Clin. Immunol. 115, 617–622 (2005).
Rouvinen, J. et al. Probing the molecular basis of allergy. Three-dimensional structure of the bovine lipocalin allergen Bos d 2. J. Biol. Chem. 274, 2337–2343 (1999).
Rautiainen, J. et al. Molecular and crystal properties of Bos d 2, an allergenic protein of the lipocalin family. Biochem. Biophys. Res. Comm. 247, 746–750 (1998).
Böcksei, Z. et al. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature 360, 186–188 (1992).
Chaudhuri, B. N. et al. The structures of α2u-globulin and its complex with a hyaline droplet inducer. Acta Cryst. D 55, 753–762 (1999).
Kamata, Y. et al. Characterization of dog allergens Can f 1 and Can f 2. 2. A comparison of Can f 1 with Can f 2 regarding their biochemical and immunological properties. Int. Arch. Allergy Immunol. 142, 301–308 (2007).
Madhurantakam, C. et al. Crystal structure of the dog lipocalin allergen Can f 2: implications for cross-reactivity to the cat allergen Fel d 4. J. Mol. Biol. 401, 68–83 (2010).
Niemi, M. H., Rytkönen-Nissinen, M., Jänis, J., Virtanen, T. & Rouvinen, J. Structural aspects of dog allergies: the crystal structure of a dog dander allergen Can f 4. Mol. Immunol. 61, 7–15 (2014).
Lacombe, M.-B. et al. Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE-reactive epitopes. J. Biol. Chem. 275, 21572–21577 (2000).
Brownlow, S. et al. Bovine β-lactoglobulin at 1.8 Å resolution – still an enigmatic lipocalin. Structure 4, 481–495 (1996).
Niemi, M. et al. Molecular interactions between a recombinant IgE antibody and the β-lactoglobulin allergen. Structure 15, 1413–1421 (2007).
Tan, Y. W. et al. Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes. J. Biol. Chem. 284, 3148–3157 (2009).
Hilger, C., Kuehn, A. & Hentges, F. Animal lipocalin allergens. Curr. Allergy Asthma Rep. 12, 438–447 (2012).
Dayhoff, J. E., Shoemaker, B. A., Bryant, S. H. & Panchenko, A. R. Evolution of protein binding modes in homooligomers. J. Mol. Biol. 395, 860–870 (2010).
Invernizzi, H. et al. Comparison of bovine and porcine β-lactoglobulin: a mass spectrometric analysis. J. Mass Spectrom. 41, 717–727 (2006).
Sakurai, K. & Goto, Y. Manipulating monomer-dimer equilibrium of bovine β-lactoglobulin by amino acid substitution. J. Biol. Chem. 28, 25735–25740 (2002).
Bello, M., Perez-Hernandez, G., Fernandez-Velasco, D. J., Arreguin-Espinosa, R. & Garcia-Hernandez, E. Energetics of protein homodimerization: effects of water sequestering on the formation of β-lactoglobulin dimer. Proteins 70, 1475–1487 (2008).
Bello, M., Portillo-Tellez, M. d. C. & Garcia-Hernandez, E. Energetics of ligand recognition and self-association of bovine β-lactoglobulin: differences between variants A and B. Biochemistry 50, 151–161 (2011).
Liu, J. & Konermann, L. Protein–protein binding affinities in solution determined by electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 22, 408–417 (2011).
Mercadante, D. et al. Bovine β-lactoglobulin is dimeric under imitative physiological conditions: dissociation equilibrium and rate constants over the pH range of 2.5–7.5. Biophys. J. 103, 303–312 (2012).
Gottschalk, M., Nilsson, H., Roos, H. & Halle, B. Protein self-association in solution: The bovine β-lactoglobulin dimer and octamer. Protein Sci. 12, 2404–2411 (2003).
Nooren, I. M. A. & Thornton, J. M. Structural characterisation and functional significance of transient protein–protein interactions. J. Mol. Biol. 325, 991–1018 (2003).
Dafforn, T. R. So how do you know you have a macromolecular complex? Acta Cryst. D 63, 17–25 (2007).
Gregoire, C. et al. Crystallization and preliminary crystallographic analysis of the major horse allergen Equ c 1. Acta Cryst. D 55, 880–882 (1999).
Davies, J. M., Platts-Mills, T. A. & Aalberse, R. C. The enigma of IgE+ B-cell memory in human subjects. J. Allergy Clin. Immunol. 131, 972–976 (2013).
Kuriyan, J. & Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 (2007).
Lin, W.-C. et al. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. 111, 2996–3001 (2014).
Kabsch, W. J. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constant. J. Appl. Cryst. 26, 795–800 (1993).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Winn, M. D. et al. Overview on the CCP4 suite and current developments. Acta Cryst. D 67, 235–242 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Cryst. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D 66, 213–221 (2010).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We acknowledge the European Synchrotron Radiation Facility for provision of its synchrotron radiation facilities and we would like to thank the staff of beamline ID-29 for their assistance in data-collection. This work was supported by the Academy of Finland (project 137988).
Author information
Authors and Affiliations
Contributions
M.R.-N. performed protein production and purification, M.N. and J.R. performed the structure determination and structure analysis. M.N., I.M. and J.J. conducted the mass spectrometric analysis; M.N., M.R.-N., J.J., T.V. and J.R. contributed to discussions and revised the manucript. J.R., M.N. and J.J. wrote the manuscript.
Ethics declarations
Competing interests
M.N., J.J. and J.R. are shareholders of Desentum Oy. The rest of the authors declare that they have no relevant conflicts of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Niemi, M., Rytkönen-Nissinen, M., Miettinen, I. et al. Dimerization of lipocalin allergens. Sci Rep 5, 13841 (2015). https://doi.org/10.1038/srep13841
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep13841
This article is cited by
-
Role of Small Molecule Ligands in IgE-Mediated Allergy
Current Allergy and Asthma Reports (2023)
-
Impact of oligomerization on the allergenicity of allergens
Clinical and Molecular Allergy (2022)
-
Serum amyloid A is a soluble pattern recognition receptor that drives type 2 immunity
Nature Immunology (2020)
-
Development of hypoallergenic variants of the major horse allergen Equ c 1 for immunotherapy by rational structure based engineering
Scientific Reports (2019)
-
Substrate prediction of Ixodes ricinus salivary lipocalins differentially expressed during Borrelia afzelii infection
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.