Abstract
RNA-binding proteins (RBPs) play an important role in plant host-microbe interactions. In this study, we show that the plant RBP known as FPA, which regulates 3′-end mRNA polyadenylation, negatively regulates basal resistance to bacterial pathogen Pseudomonas syringae in Arabidopsis. A custom microarray analysis reveals that flg22, a peptide derived from bacterial flagellins, induces expression of alternatively polyadenylated isoforms of mRNA encoding the defence-related transcriptional repressor ETHYLENE RESPONSE FACTOR 4 (ERF4), which is regulated by FPA. Flg22 induces expression of a novel isoform of ERF4 that lacks the ERF-associated amphiphilic repression (EAR) motif, while FPA inhibits this induction. The EAR-lacking isoform of ERF4 acts as a transcriptional activator in vivo and suppresses the flg22-dependent reactive oxygen species burst. We propose that FPA controls use of proximal polyadenylation sites of ERF4, which quantitatively limit the defence response output.
Similar content being viewed by others
Introduction
Plants are non-hosts to most pathogens due to pathogen associated molecular patterns (PAMP) triggered immunity (PTI). PTI is elicited through the PAMPs by extracellular pattern recognition receptors (PRRs) at the cell surface, resulting in widespread transcriptional reprogramming and broad-spectrum defence against potential pathogens. A well characterised PAMP-PRR interaction is the detection of the conserved 22-amino-acid peptide from bacteria, flg22, by the leucine-rich receptor kinase FLAGELLIN SENSITIVE2 (FLS2)1,2.
Up to now, characterisation of the plant immune response has focused mostly on defining pathogen perception followed by activation of signalling cascades and rapid changes in transcriptional programmes. However, mechanism(s) by which the plant defence response is fine-tuned to avoid associated fitness costs is less well known. Emerging evidence indicates that regulation at the RNA level comprises another layer of regulation for both pathogen virulence and plant defence3,4. For instance, the RNA-binding protein (RBP) RBP35 is required for full virulence and development in the rice blast pathogen Magnaporthe grisea5 whereas plant RBPs RBP-DR1 (RNA-BINDING PROTEIN-DEFENSE RELATED 1) and GRP7 (GLYCINE-RICH RNA-BINDING PROTEIN 7), positively regulate resistance to bacteria in Arabidopsis6,7,8,9.
Alternative polyadenylation (APA) and splicing in plants are widespread: approximately 60–70% of all Arabidopsis genes are known to contain multiple poly(A) sites10,11,12. Widespread changes in APA are associated with mammalian cancerous cells13,14 and differential poly(A) site choice has been observed in plants upon developmental changes or in response to salicylic acid treatment11. The Arabidopsis RBP FPA, a Spen family protein containing three RNA recognition motifs (RRMs), has been shown to regulate the 3′ end site choice at diverse mRNAs. The precise mechanism by which FPA regulates RNA 3′ end formation is unclear since FPA is not a known component of the conserved cleavage or polyadenylation apparatus15,16. FPA enables the transition to flowering by repressing FLOWERING LOCUS C expression17 but a role for this protein in a stress or plant defence response has not been shown.
Here, we show that FPA negatively regulates PTI responses such as the flg22-triggered ROS burst and bacterial accumulation in Arabidopsis. To see whether the role of FPA as a modulator of 3′ end RNA cleavage site choice contributes to the defence response mediated by FPA, we first searched for defence-related genes showing differential 3′ end RNA processing profiles in PTI. One such candidate was ETHYLENE RESPONSE FACTOR 4 (ERF4). ERFs are plant-specific transcription factors characterized by an APETALA2/ERF domain, which binds to DNA cis elements such as GCC boxes in target gene promoters18 to regulate diverse biological processes. In Arabidopsis, the majority of the ERF proteins are transcriptional activators, but at least eight ERFs, including ERF4, possess an EAR (Ethylene-responsive element binding factor-associated Amphiphilic Repression) motif that facilitates transcriptional repression19.
Here, we show that FPA regulates ERF4 poly(A) site choice and importantly, that ERF4 APA determines ERF4 activity. We report that ERF4 alternative polyadenylation is induced by the PAMP flg22 and that FPA inhibits this induction. Our results reveal a novel function for FPA in plant defense to bacteria and reveal a link between APA and plant immunity.
Results
The RNA-binding domain protein FPA negatively regulates PTI
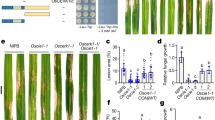
Arabidopsis mutants defective in the RNA-binding domain protein FPA exhibit RNA 3′ misregulation15. Given that several RBPs have been shown to affect host-microbe interactions, we analysed fpa loss-of- function and FPA overexpressor lines for their response to biotic stress. We first asked whether these lines exhibit altered release of reactive oxygen species (ROS), which act as signalling messengers in response to diverse biotic and abiotic stimuli. fpa mutants displayed an enhanced flg22-triggered ROS burst, whereas this was diminished in plants overexpressing FPA (Fig. 1A). Next, we asked if fpa mutants showed an altered response to the bacterial pathogen Pseudomonas syringae, as an enhanced transient ROS burst has previously been reported to correlate with increased PTI20,21. Following either spray or syringe inoculation of P. syringae isolates, 35S::FPA leaves accumulated 30- or 7-fold more bacteria than WT, respectively (Fig. 1B), suggesting that FPA can modulate basal plant defense in Arabidopsis.
FPA negatively regulates plant immunity.
(A) Oxidative burst elicited by flg22 (1 μM) in Col-0, fls2, 35S::FPA, fpa-7 and fpa-8 seedlings. Mean data with standard error from 3 biological replicates are shown. (B) Number of bacteria extracted from leaves of plants 3 days post-spray inoculation with PstDC3000ΔavrPtoΔavrPtoB (OD600 = 0.2; left panel) or 3 days post-syringe infiltration with PstDC3000 (OD600 = 0.002; right panel). Data shown are the mean colony forming units (CFU)/cm2 extracted from leaf discs from five plants per genotype. Standard errors are shown. Asterisks indicate a difference from Col-0 (P < 0.05).
Flg22 sensing triggers transcription of alternatively polyadenylated forms of the transcription factor ERF4
Since it is known that FPA modulates the 3′ end cleavage of RNAs, we searched for novel alternatively polyadenylated transcripts associated with PTI by examining the response of Arabidopsis seedlings to flg22, a well-characterized PAMP. For this purpose, we used a custom cDNA microarray based on TAIR8 annotation, containing approximately 6000 additional unannotated intergenic regions22. We compared the transcript profile of Arabidopsis wild type Col-0 (WT) seedlings with that of the flg22-insensitive receptor mutant fls2 at 0, 15, 30 and 60 min after flg22 treatment. As expected, previously characterised PTI marker genes such as WRKY29, WRKY11 and MITOGEN-ACTIVATED PROTEIN KINASE 323 were induced by flg22 in Col-0 but not fls2 (Table S1). Two hundred and twenty-four probes corresponding to unannotated intergenic regions detected differentially expressed signals with >2 fold in Col-0 and <2 fold in fls2 in at least two time points compared to the mock treatment. Among them, forty-two probes detecting high flg22 induction were selected and differential induction was quantified using quantitative RT-PCR (RT-qPCR) (Table S2). One of the most strongly induced signals corresponded to the probe ATRIKEN28815, which was chosen for further study (Fig. 2A). Rapid amplification of cDNA ends (RACE) was used to detect the full-length cDNA corresponding to ATRIKEN28815 and identified two transcripts (2445 nt and 1121 nt in length), which defined previously undetected alternatively polyadenylated isoforms of ERF4 mRNA (Figs. 2B and S1).
Alternatively polyadenylated and spliced isoforms of ERF4 are rapidly and transiently induced by flg22 treatment.
(A) Quantitative RT-PCR (RT-qPCR) validation of ATRIKEN28815 expression in Col-0 (filled circle) and fls2 (open circle) seedlings following treatment with 1 μM flg22. Data show the mean and standard error of two biological replicates. Asterisks indicate differences between Col-0 and fls2 (P < 0.05). (B) i) Schematic diagram of a 7.5-kB genomic fragment, indicating annotated ERF4 (ERF4-R), ATRIKEN28815 probe and flanking gene locations. Full-length cDNAs corresponding to ERF4-A and ERF4-IR are indicated below the annotated genes. Thick arrowheads denote exons, thicker lines denote UTRs and thin lines denote introns. ii) Primers used for transcriptional analyses. Conventional RT-PCR primers used in figure (D) are indicated by arrowheads, RT-qPCR amplicons used in (E) are indicated by thick lines and thin lines indicate the intron present in the ERF4-A amplicon. (C) The predicted ERF4-A coding region lacks the EAR motif present in both ERF4-R and ERF4-IR. (D) Conventional RT-PCR using primers P1 and P3 (top panel; 35 cycles) or P1 and P2 (bottom panel; 30 cycles) shows that the distally polyadenylated intron-retaining isoform ERF4-IR is rapidly induced within 15 min, followed by induction of the distally polyadenylated spliced isoform ERF4-A after 30 min. (E) RT-qPCR analyses of ERF4 isoforms in Col-0 or fls2 seedlings following treatment with water (0 time point) or 1 μM flg22. Asterisks indicate differences between Col-0 and fls2 (P < 0.05). Respective amplicons are shown in (B).
The 5′ end of these newly detected ERF4 mRNA isoforms map to the 5′ untranslated region (UTR) of ERF4, while the 3′ end maps to an intergenic region 1304 nt downstream of ERF4 (as annotated in the most recent Arabidopsis genome release, TAIR10) (Fig. 2B). These experiments showed that the two distally polyadenylated ERF4 transcripts differ by the presence of an intron, the excision of which removes a sequence encoding the ERF4 EAR motif (Figs. 2B, 2C, S1). Hereafter, we refer to the annotated ERF4 as ERF4-R (for repressor), the new longer transcript as ERF4-IR (for intron retention) and the new shorter transcript as ERF4-A (for activator, see below; Fig. 2B).
To quantify the ERF4 APA levels, we designed RT-qPCR primers to specifically amplify the two alternatively polyadenylated ERF4 isoforms, ERF4-A and ERF4-IR. Since ERF4-IR and ERF4-R sequences overlap, ERF4-R primers amplify both ERF4-R and ERF4-IR isoforms (Fig. 2B). We characterised in detail expression of ERF4 isoforms in response to flg22. ERF4-R was detectable by RT-qPCR with or without flg22 treatment in both fls2 and Col-0 seedlings, but ERF4-IR and ERF4-A were only observed 15–30 min or 30–60 min, respectively, after flg22 treatment of WT seedlings (Fig. 2D). This RNA processing response was transient, as ERF4 APA transcript levels returned to almost basal levels by 3–6 h post-flg22 treatment (Fig. 2E). ERF4-R was induced slightly in fls2, but neither ERF4-A or ERF4-IR expression was induced in fls2 at any time point, demonstrating the requirement of flg22 sensing for the induction of ERF4 APA. To determine if the newly identified alternatively polyadenylated ERF4 isoforms were also found in response to other biotic stress-related treatments, we tested expression of ERF4-R, ERF4-IR and ERF4-A in response to defence hormones salicylic acid or methyl jasmonate (MeJA). Although ERF4-A was strongly induced by flg22, neither SA nor MeJA significantly altered ERF4-A expression (Fig. S2), suggesting that ERF4 APA is a specific response that occurs in the early stage of PTI.
FPA affects ERF4 RNA procesing
Using direct RNA sequencing (DRS), we detected an increased abundance of distally polyadenylated transcripts corresponding to ERF4-A and ERF4-IR in fpa-7 mutants, suggesting that FPA negatively regulates ERF4 APA (Fig. 3A). The 3′ end of the ERF4 APA isoforms is cleaved in intergenic sequence with upstream canonical poly(A) cis elements (AAUAAA -19 nucleotides upstream of the cleavage site and a U-rich sequence immediately upstream of the cleavage site) previously shown to be associated with preferred cleavage sites in Arabidopsis 3′UTRs12, whereas canonical poly(A) cis elements could not be detected at the 3′ end of the annotated ERF4 isoform (Figs. S1, 3B).
Use of the ERF4 distal polyadenylation site is increased in fpa plants.
(A) Distribution of direct RNA sequencing (DRS) normalized reads at the ERF4 locus. DRS was performed on RNAs from 14-day-old seedlings of Col-0 and fca-9, fpa-7 and fpa-7 fca-9 flowering time mutants for the first experiment and of Col-0 and the fld-3 flowering time mutant for the second experiment. Representative Col-0 data for each experiment is shown above the mutant data. Exons are denoted by rectangles and UTRs by adjoining narrower rectangles. (B) Proposed designation of alternating U- and A-rich sequences at the ERF4-A or ERF4-IR downstream cleavage sites. Location of the AAUAAA and AUGUUU cis elements (AUGUUU corresponds to the point mutation in the UUGUUU motif) are displayed, as are their positions relative to the cleavage site. USE, upstream sequence element; PAS, polyA signal; Fip1, the U-rich sequence upstream of the cleavage site is the proposed Fip1-binding site56,57; DSE, downstream sequence element; black triangle, cleavage site. (C) Expression of the ERF4 amplicons indicated in Fig. 2A in Col-0, 35S::FPA, fpa-7 and fpa-8. Data represent the mean and standard error of three biological replicates. The asterisk indicates a difference from Col-0 (P < 0.05).
Importantly, we found that the alternatively polyadenylated isoforms ERF4-A and ERF4-IR were upregulated in both fpa mutant alleles relative to Col-0, with ERF4-A expressed >60 fold more in independent fpa mutant alleles compared to WT plants. In contrast, transgenic plants overexpressing FPA exhibited reduced ERF4-A expression (6 fold lower than WT), but showed WT levels of ERF4-IR expression (Fig. 3C). Together, these data indicate that FPA negatively regulates the expression of ERF4-A. Since the Arabidopsis flowering time regulator FCA can also affect RNA 3′ end formation15, we analysed ERF4 RNA processing in plants defective in FCA or FLD, a histone demethylase which also regulates flowering time in Arabidopsis through the same pathway24. Neither fca-9 nor fld-3 exhibited altered ERF4 poly(A) site selection or ERF4-A expression relative to WT plants. Furthermore, ERF4 poly(A) site selection was similar in fpa-7 fca-9 double and fpa-7 single mutants, revealing that FCA does not act redundantly with FPA in ERF4 APA control and that late flowering and elevated FLC levels do not indirectly alter ERF4 APA (Figs. 3A, S3).
APA of ERF4 generates new functional specificities
We reasoned that ERF4-A might lack transcription repressor activity due to loss of the EAR motif. To test this hypothesis, we analysed expression of the luciferase (LUC)-encoding reporter gene fused to a GCC box promoter using a transient transcriptional activity assay. When introduced by bombardment into Arabidopsis cells the ERF4-R coding region reduced the expression of the reporter gene >2-fold, whereas expression of the ERF4-A coding region activated the expression of the reporter gene >7-fold (Fig. 4A). These data indicate that deletion of the EAR motif, caused by alternative processing of ERF4 pre-mRNA in response to flg22 perception, switches ERF4 from being a transcriptional repressor to an activator.
Overexpression of ERF4-R or ERF4-A produce contrasting phenotypes.
(A) Relative luciferase activities after cobombardment of Arabidopsis suspension cells with the GAL4GCC-LUC reporter gene. Data represent the mean and standard error of three biological replicates. Asterisks indicate differences from the empty vector control (P < 0.05). (B) Representative 6-week-old plants grown under long day conditions showing the morphological effects of ERF4-A and ERF4-R ORF overexpression.
To assess potential biological roles of ERF4-A and ERF4-R in planta, we generated transgenic Arabidopsis lines stably overexpressing the same ERF4 isoforms as those used in the transient assays. The multiple lines highly expressing ERF4-A under the control of the 35S promoter were larger than WT plants, whereas the majority of the 35S::ERF4-R::nos transformants were stunted when grown under long day growth conditions (Fig. 4B). The contrasting phenotypes of these lines are consistent with the idea that alternatively polyadenylated ERF4 isoforms encode proteins with distinct in vivo functions.
To understand the individual biological roles of ERF4-A and ERF4-R in plant defence, we transformed the T-DNA insertion line erf425 with either the ERF4-R or ERF4-A isoform under the control of the native ERF4 promoter (referred to as erf4/ERF4-R and erf4/ERF4-A; Figs. 5A and S4). We first asked whether these lines exhibit altered release of flg22-dependent ROS. Independent erf4/ERF4-A and 35S::ERF4-A::nos lines exhibited reduced ROS bursts relative to WT and erf4 whereas independent erf4/ERF4-R and 35S::ERF4-R::nos lines exhibited enhanced ROS bursts relative to WT (Fig. 5B).
ERF4 isoforms differentially regulate both flg22-triggered ROS burst and PDF1.2 expression.
(A) RT-PCR showing the presence or absence of ERF4 isoforms 30 min after 1 μM flg22 treatment of Col-0, erf4, erf4/ERF4-A and erf4/ERF4-R seedlings. (B) The oxidative burst elicited by 1 μM flg22 in representative ERF4 isoform complementation (left) and overexpression (right) lines. Data represent the mean and standard error of three biological replicates. (C). Expression of PDF1.2 in 14-day-old seedlings of the indicated genotypes. Data represent the mean and standard error of three biological replicates.
We next asked how the expression of PDF1.2, which encodes a defensin involved in the jasmonate defence signalling pathway is affected by different ERF4 isoforms. ERF4-R was previously shown to be a negative regulator of PDF1.225,26. Indeed, 35S::ERF4-R::nos and erf4/ERF4-R seedlings showed reduced basal PDF1.2 expression relative to WT whereas erf4 and 35S::ERF4-A::nos lines exhibited enhanced basal PDF1.2 expression relative to WT (Fig. 5C). These data reveal that ERF4-A and ERF4-R isoforms have opposing roles in regulating both the ROS burst and PDF1.2 expression. However, erf4, as well as complementation and overexpression lines, responded similarly to WT plants when inoculated with the bacterial pathogen P. syringae under our experimental conditions (Fig. S5).
Flg22-triggered induction of ERF4-A is inhibited by FPA
We next examined whether the FPA- and flg22-dependent control of ERF4 APA might be mediated through a single signalling pathway. Basal expression of ERF4 APA transcripts was increased in independent fpa mutant alleles, whereas 35S::FPA plants exhibited a reduction in ERF4 APA transcripts relative to WT (Figs. 3C, 6A). Since flg22 and FPA differentially modulate poly(A) site usage at ERF4, we speculated that disruption of FPA function might account for increased ERF4 readthrough upon flg22 treatment. In such a scenario, flg22-triggered induction of ERF4 APA would be abolished in fpa mutants. However, on the contrary, ERF4 APA isoforms increased both in WT and fpa mutants after flg22 treatment (Fig. 6A). These data indicate the possibility that genetically distinct pathways converge to regulate ERF4 RNA processing. ERF4-R remained the predominant ERF4 isoform in all genotypes, either before or after flg22 treatment (Figs. 2E, 6A), suggesting that increased ERF4 APA isoforms in fpa mutants or in flg22 treated plants cannot be explained solely as a consequence of a binary switch in poly(A) site use. Together, these data suggest that flg22 promotes use of the canonical distal poly(A) site, possibly by inhibiting use of the proximal poly(A) site to facilitate readthrough, resulting in increased expression of the ERF4-A isoform. On the other hand, FPA, at least partially independently of flg22, may promote use of the proximal poly(A) site, resulting in ERF4-R expression and inhibiting readthrough (Fig. 6B).
FPA inhibits flg22-triggered induction of ERF4-A.
(A) Expression of the ERF4 amplicons indicated in Fig. 2B in Col-0, fls2, 35S::FPA, fpa-7 and fpa-8 seedlings either untreated (left) or treated with 1 μM flg22 for 1 h (right). Data represent the mean and standard error of three biological replicates. The asterisk indicates difference from Col-0 (P < 0.05). (B) Schematic model indicating the role of flg22 and FPA in ERF4 processing. FPA promotes use of the proximal poly(A) site, inhibiting readthrough to the canonical distal poly(A) site. flg22 promotes distal poly(A) site use, possibly by inhibiting proximal poly(A) site use, allowing readthrough and subsequent splicing.
Discussion
Emerging evidence indicates that RNA processing is part of an active defense response in plants27. APA has previously been implicated as a modulator of plant immune responses mainly because mutations in genes involved in APA or RNA processing show altered defense responses and disease phenotypes7,8,28. In most cases, how poly(A) site choice is regulated during plant defense is not known. In this study, we uncovered a novel mechanism by which transcription factor activity may be regulated as a consequence of FPA mediated 3′ end RNA processing and alternative splicing.
FPA contains a RRM but the precise mechanism by which FPA controls 3′ end formation (and possibly other processes such as splicing) of RNA targets is unclear. FPA controls polyadenylation of its own transcript and of others by promoting polyadenylation at the proximal site, thereby inhibiting transcriptional readthrough to a stronger distal poly(A) cis element16. At ERF4, an identified ultimate target of FPA, intergenic distal poly(A) cis elements are canonical, suggesting a strong poly(A) site that does not depend on FPA function. Since the expression of the proximally polyadenylated isoform ERF4-R accumulated to higher levels than ERF4 APA transcripts ERF4-A or ERF4-IR, even in the absence of a functional copy of FPA, proteins with partial functional redundancy to FPA, or more generic cleavage and poly(A) complexes are likely to promote proximal poly(A) site use at ERF4. FCA has been shown to act synergistically with FPA to control 3′ ends at several loci15, however neither fca nor fld loss-of-function mutants24 exhibited altered RNA processing at ERF4.
Loss of function fpa mutants exhibit defective 3′ end formation and consequent intergenic read-through at specific loci15,16. We show here that such read-through events are not necessarily benign. Read-through is often associated with cryptic splicing events that are not normally detected in WT15. We demonstrate here that such read-through coupled with cryptic splicing can alter the coding sequence and function of the upstream gene. These otherwise unpredicted consequent changes in RNA processing therefore have generally important implications for understanding the impact of APA and defective RNA 3′ end formation on gene regulation and disease29.
In this study, we showed that APA of ERF4 is of biological relevance since it results in generation of a new ERF4 isoform that lacks the well characterized EAR repression motif of this transcription factor. Indeed, using in vivo experiments we showed that this new ERF4 isoform, named as ERF4-A, acts as a transcriptional activator. Mechanisms such as phosphorylation30 or proteolysis31 have been shown or proposed to regulate the activity of ERF proteins. Our results show that APA-mediated EAR motif deletion is a novel mechanism by which ERF transcription factor activity can be manipulated. ERF4 transcripts are highly unstable32, suggesting a high turnover of mRNA whose polyA site usage can be regulated depending on the signalling requirements of the plant. We propose that ERF4-A acts to dampen the amplitude of the ROS burst, thereby preventing a ‘runaway’ defence response.
Intriguingly, when expressed in transgenic plants, ERF4-A acts as a negative regulator of flg22-triggered ROS burst. Therefore, one important function of ERF4-A generated by flg22 might be to control ROS rapidly generated in plant cells upon perception of flg22, so that potentially detrimental effects of these important signalling (but also damaging) molecules on plant cells can be minimized. Direct downstream targets of ERF4 are still largely unknown, but PDF1.2, encoding a plant defensin with antimicrobial properties, has been shown to be repressed by ERF425. As expected, PDF1.2 was differentially expressed in transgenic plants expressing ERF4-A or ERF4-R. OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF-domain protein 59 (ORA59), ERF104 and ERF1 have been shown to bind directly to the promoter of PDF1.233,34,35, but to date, there is no experimental evidence showing that ERF4 binds directly to the PDF1.2 promoter. Therefore ERF4 may act upstream or in competition with these ERFs to fine tune plant defense. When grown under long day conditions, overexpression of ERF4-R or ERF4-A resulted in T1 stunted or healthy plants, respectively, In contrast, McGrath et al. (2005) reported that 35S::ERF4 plants exhibited a WT growth morphology. Reasons for this discrepancy may include variations in environmental growth conditions or the constructs used to transform plants. While we transformed plants with the At3g15210 exon sequence, constructs used by McGrath et al. (2005) included regions of the At3g15210 3′ and 5′ UTRs.
ERF4 interacts with several corepressor proteins including TOPLESS (TPL), TPL-related (TPR), SIN3 ASSOCIATED POLYPEPTIDE 18 and ARABIDOPSIS HISTONE DEACETYLASE 1936,37, which recruit chromatin modifying enzymes required for transcriptional suppression38,39. TOPLESS has been shown to bind directly to the EAR motif in several protein-protein interactions40,41, suggesting that the ERF4-TOPLESS interaction might be EAR-motif dependent. Given that ERF4-A and ERF4-R differ by the presence or absence of the EAR motif, EAR-motif dependent protein interactions could result in differential function or even subcellular localization of ERF4-R and ER4-A. It is also possible that ERF4 isoforms compete for the same DNA binding site or heterodimerize in planta.
Interestingly, altered APA at ERF4 in fpa could not account for the enhanced defence phenotype in fpa. Since intergenic regions downstream of several loci are upregulated in fpa15, ERF4 is likely to be one of several defence genes whose RNA processing is regulated by FPA. Furthermore, flg22-triggered modulation of APA may not be specific to ERF4. Identification of the defence-associated gene(s) or factors which are modulated by FPA and upon biotic stress requires further investigation. It is likely that APA at multiple loci in fpa could contribute quantitatively to achieve an overall positive regulation of PTI (Fig. 7). Plants are sessile organisms and are exposed to an abundance of microbes. While some microbes that come into contact with the plant are pathogenic, the majority are likely to be non-pathogenic. PTI is therefore under tight negative regulation to avoid expending energy on defence when it is not required. APA is likely to be an important process that allows the plant to rapidly expand the complexity of the transcriptome and/or proteome in response to flg22 and other stresses, but must be kept ‘under wraps’ to avoid an overreaction of the plant.
Schematic model integrating the role of FPA and ERF4 isoforms in modulating the flg22-triggered ROS burst output.
Upon flg22 treatment, the ERF4 promoter is activated and expression of genes encoding both ERF4-R and ERF4-A, which positively and negatively regulate the ROS burst, respectively, increase. FPA partially inhibits the flg22-triggered induction of ERF4-A by promoting polyadenylation at ERF4-R (Fig S6). While ERF4-A suppresses the flg22-triggered ROS burst, FPA also suppresses the ROS burst. Therefore, APA modulation at ERF4 cannot explain increased ROS burst in fpa seedlings. It is possible that ERF4 is one of multiple defence genes targeted by FPA under basal conditions or upon flg22 treatment in which altered APA, potentially leading to protein coding changes, results in the suppression of ROS production. We propose that such APA events are induced by flg22, but are inhibited by FPA to limit excess resource allocation to defense when it is not required.
In addition to its role in defence regulation, FPA also promotes the transition to flowering. Interestingly, another Arabidopsis RBP, GRP7, also promotes the transition to flowering. In contrast to FPA, which negatively regulates PTI, GRP7 positively regulates PTI42. Plants need to maintain a tradeoff between development and defense to ensure optimal reproductive outcomes while surviving biotic and other stresses43. The interplay between flowering and defence has demonstrated by several examples in other pathosystems. Known mediators of both flowering and bacterial defence are PLANT U BOX PROTEIN 13 44, HOPW1-1-INTERACTING 3 45, LEAFY46, SIZ147 and ENHANCED DOWNY MILDEW 2 48.
In summary, the findings we present here may reflect the independent regulation by FPA and other RBPs of different targets involved in defence and flowering. Alternatively, such interactions may define a facet of the integration of these crucial life history traits, thus linking flowering with immunity at the molecular level.
Methods
Plant material, growth conditions and flg22 treatment
Arabidopsis plants were grown at 22°C under a 16 h light: 8 h dark cycle (normal day length conditions) or an 8 h light: 16 h dark cycle (short day length conditions). The WT genotype used was Col-0. erf4, fpa-7, fpa-8 and 35S::FPA lines were described previously16,25,49. Seedling treatment with flg22 was performed as described previously23. Briefly, stratified Arabidopsis Col-0 and fls2 seeds were grown on plates containing 1× Murashige and Skoog medium (MS), 1% sucrose and 0.8% agar under normal day length conditions for 12 days. Seedlings were then transferred to liquid MS containing 1 μM flg22 or water for 2 days before harvesting. The flg22 peptide was purchased from Sigma and solubilized in water.
Flg22-triggered ROS burst
The flg22-triggered ROS burst was assayed as described previously50. Ten seeds of each genotype were dispensed per well, in triplicate, into a 48-well plate (NUNC). After surface sterilization, MGRL nutrient medium51 supplemented with 0.1% sucrose was added to the plate and seeds were stratified for 2 days at 4°C. After 8 days growth under normal day length conditions, liquid medium was removed, replaced with MGRL nutrient medium supplemented with 0.1% sucrose and 100 μM L012 (Sigma) and plants were then incubated in the dark for 1.5 h. L012-containing medium was then removed and replaced with MGRL nutrient supplemented with 0.1% sucrose and 1 μM flg22. ROS production was measured immediately using a Mithras LB940 microplate luminometer (Berthold Technologies). At least 10 independent T2 transformants for each complementation line and four independent T2 transformants for each overexpression line were tested.
Bacterial spray pathotest
Six-week-old plants grown under short day conditions were sprayed with a bacterial suspension containing 1 × 108 cfu ml–1 PstDC3000ΔavrPtoΔavrPtoB52 in 10 mM MgCl2 containing 0.04% Silwet L-77 (Bio Medical Science, Japan) and covered to maintain humidity. For syringe infiltration experiments, abaxial leaf surfaces were inoculated using a 1 mL needleless syringe with bacterial suspension containing 1 × 105 cfu ml−1. Three days post-inoculation, counting of leaf bacteria by serial dilution plating was performed as previously described53.
RNA isolation, microarray hybridization, RT-PCR and RT-qPCR
RNA was extracted and DNase treated using the RNeasy mini kit (Qiagen), according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg RNA using SuperScript RNA H- Reverse Transcriptase (Invitrogen) and oligo (dT) primer, according to the manufacturer's instructions. RT-PCR products were amplified in 25-μl PCR reactions containing 2 μl of first-strand cDNA, 1×PCR Buffer, 0.2 mM dNTPs, 0.5 U of Phusion DNA Polymerase (New England Biosciences) and 0.5 μM primer P1 and primer P2 or P3 (Table S3). RT-qPCR was performed using the Thunderbird SYBR qPCR mix (Toyobo, Japan) and 0.6 pmol primers (Table S1) according to the manufacturer's instructions using a Stratagene Mx3000P sequence detection system (Agilent Technologies). Quantities were determined against a standard curve using MxPro Software (Mx3000P version 4.1; Agilent Technologies) and normalized to actin. Plasmids containing equal molar ratios of ERF4-A or ERF4-IR were used to quantify ERF4-A and ERF4-IR/ERF4-R transcripts, respectively. All other genes were quantified using cDNA from Col-0 seedlings treated with flg22 for 30 min. All reactions were carried out in duplicate. RNA from each of eight samples (Col-0 or fls2: mock (0), 15, 30 and 60 min post-flg22 treatment) were reverse transcribed and labelled with Cy3 using the Quick Amp Labeling kit (Agilent), according to the manufacturer's instructions. Labelled cRNAs were then hybridized to RIKEN custom arrays as described previously22.
Rapid amplification of cDNA ends
cDNA used for 3′ and 5′ RACE was prepared from 1 μg RNA extracted from Col-0 seedlings harvested 30 min after treatment with flg22. RACE was done using the GeneRacer kit (Invitrogen), according to the manufacturer's instructions and the gene-specific primers 28815_5′, 28815_5′ nest and 28815_3′ (Table S1). Five clones were sequenced per PCR product.
Dual-luciferase assay
To examine the transcriptional function of ERF4-R and ERF4-A, we performed a transient reporter assay using the Dual-Luciferase Reporter Assay System (Promega). To make effector constructs, the coding regions of ERF4-A and ERF4-R were amplified using ERF4_atg and ERFA_stop or ERF4R_stop primers, respectively (Table S1). The resulting ERF4-R and ERF4-A products were cloned into p35SNOSG. This vector contains the CaMV 35S promoter, a translational enhancer from tobacco mosaic virus (Ω) and the GAL4GCC-LUC reporter gene. The effector, reporter and reference plasmids were introduced into Arabidopsis MM2D cell cultures by particle bombardment using a PDS-1000 particle gun (Bio-Rad). Luciferase activity was measured using a Mithras LB940 microplate luminometer (Berthold Technologies).
Generation of Arabidopsis transgenic plants
A modified Agrobacterium tumefaciens-mediated floral dip method54 was used for plant transformation. To produce plants overexpressing ERF4-R or ERF4-A open reading frames (ORFs), effector plasmids used in the dual-luciferase assay were subcloned into the pBCKK vector and transformed into Col-0 plants. The growth morphology of 43 35S::ERF4-R::nos T1 and 50 35S::ERF4-A::nos T1 plants was assessed visually. To express ERF4 isoforms under the native promoter in the erf4 background, full-length cDNAs corresponding to ERF4 isoforms were amplified from cDNA using the primers ERF4_start and ERF4–3′_R (ERF4-R isoform) or ERF4_start and ERF4-3′_F (ERF4-A isoforms) and then fused to the native (−2401 nt) promoter amplified from gDNA using primers ERF4prom_F and ERF4prom_R (Table S1). Amplification products were cloned into pENTR (Invitrogen), subcloned into the promoterless pGWB1 vector55 and then transformed into erf4 plants.
References
Zipfel, C. et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767 (2004).
Gomez-Gomez, L. & Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5, 1003–1011 (2000).
Ambrosone, A., Costa, A., Leone, A. & Grillo, S. Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Science 182, 12–18 (2012).
Staiger, D., Korneli, C., Lummer, M. & Navarro, L. Emerging role for RNA-based regulation in plant immunity. New Phytol 197, 394–404 (2013).
Franceschetti, M. et al. Fungal virulence and development is regulated by alternative pre-mRNA 3′ end processing in Magnaporthe oryzae. PLoS Pathog 7, e1002441, 10.1371/journal.ppat.1002441 (2011).
Jeong, B. R. et al. Structure function analysis of an ADP-ribosyltransferase type III effector and its RNA-binding target in plant immunity. Journal of Biological Chemistry 286, 43272–43281 (2011).
Fu, Z. Q. et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447, 284–U281 (2007).
Lee, H. J. et al. Different roles of glycine-rich RNA-binding protein 7 in plant defense against Pectobacterium carotovorum, Botrytis cinerea and tobacco mosaic viruses. Plant Physiol Bioch 60, 46–52 (2012).
Qi, Y. et al. A putative RNA-binding protein positively regulates salicylic acid–mediated immunity in Arabidopsis. Mol. Plant-Microbe Interact. 23, 1573–1583 (2010).
Xing, D. & Li, Q. Q. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdisciplinary Reviews: RNA 2, 445–458 (2011).
Shen, Y. et al. Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing. Genome Research 21, 1478–1486 (2011).
Sherstnev, A. et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol 19, 845–852 (2012).
Mayr, C. & Bartel, D. P. Widespread Shortening of 3′ UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell 138, 673–684 (2009).
Yoon, O. K., Hsu, T. Y., Im, J. H. & Brem, R. B. Genetics and regulatory impact of alternative polyadenylation in human β-Lymphoblastoid Cells. Plos Genetics 8, ARTN e1002882DOI 10.1371 (2012).
Sonmez, C. et al. RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proceedings of the National Academy of Sciences 108, 8508–8513 (2011).
Hornyik, C., Terzi, L. C. & Simpson, G. G. The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA. Developmental Cell 18, 203–213 (2010).
Koornneef, M., Hanhart, C. J. & Vanderveen, J. H. A Genetic and Physiological Analysis of Late Flowering Mutants in Arabidopsis-Thaliana. Mol Gen Genet 229, 57–66 (1991).
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H. & Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box mediated gene expression. Plant Cell 12, 393–404 (2000).
Nakano, T., Suzuki, K., Fujimura, T. & Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology 140, 411–432 (2006).
He, P., Shan, L. & Sheen, J. Elicitation and suppression of microbe-associated molecular pattern-triggered immunity in plant–microbe interactions. Cellular Microbiology 9, 1385–1396 (2007).
Gomez-Gomez, L. & Boller, T. Flagellin perception: a paradigm for innate immunity. Trends in Plant Science 7, 251–256 (2002).
Hanada, K. et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proceedings of the National Academy of Sciences 110, 2395–2400 (2013).
Navarro, L. et al. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128 (2004).
Liu, F. Q. et al. The Arabidopsis RNA-Binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28, 398–407 (2007).
McGrath, K. C. et al. Repressor and activator type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiology 139, 949–959 (2005).
Yang, Z., Tian, L. N., Latoszek-Green, M., Brown, D. & Wu, K. Q. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology 58, 585–596 (2005).
Staiger, D., Korneli, C., Lummer, M. & Navarro, L. Emerging role for RNA-based regulation in plant immunity. New Phytol 197, 394–404 (2012).
Palma, K. et al. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes & Development 21, 1484–1493 (2007).
Di Giammartino, D. C. et al. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43(6), 853–866 (2011).
Song, C. P. et al. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396 (2005).
Kazan, K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends in Plant Science 11, 109–112 (2006).
Gutierrez, R. A., Ewing, R. M., Cherry, J. M. & Green, P. J. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proceedings of the National Academy of Sciences of the United States of America 99, 11513–11518 (2002).
Zarei, A. et al. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Molecular Biology 75, 321–331 (2011).
Pre, M. et al. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147, 1347–1357 (2008).
Bethke, G. et al. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proceedings of the National Academy of Sciences 106, 8067–8072 (2009).
Causier, B., Ashworth, M., Guo, W. & Davies, B. The TOPLESS Interactome: A Framework for Gene Repression in Arabidopsis. Plant Physiology 158, 423–38 (2011).
Song, C. P. & Galbraith, D. W. AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Molecular Biology 60, 241–257 (2006).
Long, J. A., Ohno, C., Smith, Z. R. & Meyerowitz, E. M. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 (2006).
Kagale, S. & Rozwadowski, K. EAR motif-mediated transcriptional repression in plants An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146 (2011).
Pauwels, L. et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–U169 (2010).
Szemenyei, H., Hannon, M. & Long, J. A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008).
Streitner, C. et al. The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. The Plant Journal 56, 239–250 (2008).
Alcázar, R., Reymond, M., Schmitz, G. & de Meaux, J. Genetic and evolutionary perspectives on the interplay between plant immunity and development. Current Opinion in Plant Biology 14, 378–384 (2011)
Li, W. et al. The U-Box/ARM E3 Ligase PUB13 Regulates Cell Death, Defense and Flowering Time in Arabidopsis. Plant Physiology 159, 239–250 (2012).
Wang, G. et al. Multiple Roles of WIN3 in Regulating Disease Resistance, Cell Death and Flowering Time in Arabidopsis. Plant Physiology 156, 1508–1519.
Winter, C. M. et al. LEAFY Target Genes Reveal Floral Regulatory Logic, cis Motifs and a Link to Biotic Stimulus Response. Developmental Cell 20, 430–443 (2011).
Jin, J. B. et al. The SUMO E3 ligase, AtS1Z1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant Journal 53, 530–540 (2008).
Tsuchiya, T. & Eulgem, T. The Arabidopsis defense component EDM2 affects the floral transition in an FLC-dependent manner. Plant Journal 62(3), 518–528 (2010).
Baurle, I., Smith, L., Baulcombe, D. C. & Dean, C. Widespread Role for the Flowering-Time Regulators FCA and FPA in RNA-Mediated Chromatin Silencing. Science 318, 109–112 (2007).
Albert, P., Miya, A., Hiratsuka, K., Kawakami, N. & Shibuya, N. A high-throughput evaluation system for Arabidopsis mutants for defense signaling. Plant Biotechnology 23, 459–466 (2006).
Fujiwara, T., Hirai, M. Y., Chino, M., Komeda, Y. & Naito, S. Effects of Sulfur Nutrition on Expression of the Soybean Seed Storage Protein Genes in Transgenic Petunia. Plant Physiol. 99, 263–268 (1992).
Shan, L. B. et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host & Microbe 4, 17–27 (2008).
Katagiri, F., Thilmony, R. & He, S. in The Arabidopsis Book (eds Somerville C. R., & Meyerowitz E. M.) American Society of Plant Biologists, (2002).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743 (1998).
Nakagawa, T. et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering 104, 34–41 (2007).
Jan, C. H., Friedman, R. C., Ruby, J. G. & Bartel, D. P. Formation, regulation and evolution of Caenorhabditis elegans 3′ UTRs. Nature 469, 97–U114 (2011).
Kaufmann, I., Martin, G., Friedlein, A., Langen, H. & Keller, W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. Embo Journal 23, 616–626 (2004).
Acknowledgements
This work was partially supported by the grant-in-aid (KAKENHI no. 24228008 to K.S.), the Program for Promotion of Basic and Applied Researchers for Innovations in Bio-oriented Industry (BRAIN to K.H. and M.H.), the Scottish Government and BBSRC grants (BB/J00247X/1 and BB/H002286/1 to G.S) and by RIKEN Foreign Postdoctoral Researcher (FPR) and CSIRO OCE (Office of the Chief Executive) postdoctoral fellowships to R.L. We thank Kaori Takizawa and Akiko Ueno for technical assistance.
Author information
Authors and Affiliations
Contributions
R.L., G.S., K.S. and K.S.* designed research; R.L., A.I., T.G., C.D. and M.H. performed research, K.H. and M.M. contributed new reagents/analytic tools; R.L., A.S., C.D., G.J.B. analyzed data; and R.L., K.K., K.S.* and G.S. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Tables 1, 2 and 3
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Lyons, R., Iwase, A., Gänsewig, T. et al. The RNA-binding protein FPA regulates flg22-triggered defense responses and transcription factor activity by alternative polyadenylation. Sci Rep 3, 2866 (2013). https://doi.org/10.1038/srep02866
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02866
This article is cited by
-
The nucleoporin NUP160 and NUP96 regulate nucleocytoplasmic export of mRNAs and participate in ethylene signaling and response in Arabidopsis
Plant Cell Reports (2023)
-
A chimeric AtERF4 repressor modulates pleiotropic aspects of plant growth and abiotic stress tolerance in transgenic Arabidopsis
Plant Growth Regulation (2022)
-
Arabidopsis CPR5 plays a role in regulating nucleocytoplasmic transport of mRNAs in ethylene signaling pathway
Plant Cell Reports (2022)
-
Leaf senescence: progression, regulation, and application
Molecular Horticulture (2021)
-
Analysing the genetic architecture of clubroot resistance variation in Brassica napus by associative transcriptomics
Molecular Breeding (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.