Abstract
The biosynthetic pathway for the rare compatible solute mannosylglucosylglycerate (MGG) accumulated by Rhodopirellula baltica, a marine member of the phylum Planctomycetes, has been elucidated. Like one of the pathways used in the thermophilic bacterium Petrotoga mobilis, it has genes coding for glucosyl-3-phosphoglycerate synthase (GpgS) and mannosylglucosyl-3-phosphoglycerate (MGPG) synthase (MggA). However, unlike Ptg. mobilis, the mesophilic R. baltica uses a novel and very specific MGPG phosphatase (MggB). It also lacks a key enzyme of the alternative pathway in Ptg. mobilis – the mannosylglucosylglycerate synthase (MggS) that catalyses the condensation of glucosylglycerate with GDP-mannose to produce MGG. The R. baltica enzymes GpgS, MggA and MggB were expressed in E. coli and characterized in terms of kinetic parameters, substrate specificity, temperature and pH dependence. This is the first characterization of genes and enzymes for the synthesis of compatible solutes in the phylum Planctomycetes and for the synthesis of MGG in a mesophile.
Similar content being viewed by others
Introduction
Planctomycetes form a deep branching bacterial phylum comprising budding, peptidoglycan-less organisms with unusual features of enormous importance for understanding cell biology and the evolution of cell organization. All members of this phylum share a unique cell plan in which the cell cytoplasm is divided into compartments by one or more membranes, including a major cell compartment containing the nucleoid1. Planctomycetes are found in a broad range of environments, including terrestrial soil and aquatic systems of different ionic composition, e.g. hypersaline, marine water, oceanic abyssal sediments and freshwater2.
Saline environments represent a challenge to a microorganism's survival due to the low water activity and to the high concentrations of inorganic ions that are toxic to cellular metabolism. However, most microorganisms respond to osmotic stress by accumulating low molecular weight soluble metabolites, termed compatible solutes, either by uptake from the medium or by de novo synthesis, many of which accumulate to very high levels3. The large diversity of compatible solutes falls into a few major chemical categories such as polyols and derivatives, sugars and derivatives, amino acids and derivatives, betaines and ectoines. While some are widely distributed in nature others seem to be exclusively present in specific groups of organisms3,4.
A different and unique compatible solute derived from glucosylglycerate (GG) was recently identified in deep-branching bacteria Petrotoga miotherma and Ptg. mobilis from the order Thermotogales, and characterized as mannosyl-(1,2)-glucosylglycerate (MGG)5,6. In Ptg. miotherma, MGG was the principal compatible solute along with minor amounts of glutamate and proline, although MGG only seemed to play a role in the organism's adaptation to suboptimal salinities5. On the other hand, Ptg. mobilis accumulated MGG during growth at supra-optimal salinities and temperatures6.
MGG, unexpectedly, was recently shown to accumulate in the mesophilic Rhodopirellula baltica7 (SH 1T)7, a pink-colored marine aerobic heterotrophic representative of the Planctomycetes8. The identification of MGG in this deep-rooted organism1,7 prompted us to examine the pathway for its synthesis.
Two independent biosynthetic pathways for the synthesis of MGG have been elucidated in Ptg. mobilis6. One, the phosphorylating three-step pathway, where UDP-glucose and D-3-phosphoglycerate (3-PGA) formed the intermediate glucosyl-3-phosphoglycerate (GPG) by the action of a glucosyl-3-phosphoglycerate synthase (GpgS). A mannosylglucosyl-3-phosphoglycerate synthase, designated MggA, uses the product of GpgS (GPG), as well as GDP-mannose, to produce mannosylglucosyl-3-phosphoglycerate (MGPG), the phosphorylated precursor of MGG, which is then dephosphorylated by a putative mannosylglucosyl-3-phosphoglycerate phosphatase (MggB) to MGG. The alternative two-step nonphosphorylating pathway, involves the direct condensation of ADP-glucose and D-glycerate into GG by a glucosylglycerate synthase (GgS), followed by the conversion of GDP-mannose and GG into MGG by a mannosylglucosylglycerate synthase (MggS).
The R. baltica GpgS (rb1005) and MggA (rb5546) genes were expressed in E. coli and the recombinant enzymes were purified and characterized. However, a gene for the MggB phosphatase was absent from R. baltica genome. Nevertheless, two genes encoding a putative MggS and an unknown hydrolase (rb12347) were detected in R. baltica genome. Herein, we show that the hydrolase is in fact the first described phosphatase specific for the MGPG dephosphorylation, MggB, with a rather scarce distribution among the sequenced genome databases.
To understand the role of MGG at low temperatures and the regulatory mechanisms involved in the adaptation of R. baltica to salt stress, elucidation of the metabolic pathway is essential. In this study, we examine the synthesis of MGG through the phosphorylating pathway from D-3-phosphoglycerate and ADP-glucose to the final compatible solute, in cell extracts and by functional characterization of recombinant enzymes.
Results
Identification, functional overexpression and purification of the recombinant GpgS, MggA and MggB
BLAST searches in the R. baltica SH1 genome database with Ptg. mobilis, Persephonella marina and Methanococcoides burtonii GpgS and with Ptg. mobilis MggA and MggS6,9,10, resulted in the identification of three open reading frames with homology to GpgS, MggA and MggS, which were then functionally expressed in E. coli. An operon-like structure containing the putative mggS was identified containing two additional genes encoding for a putative phosphatase (mggB) and a putative sucrose phosphorylase (spasE).
Expression of R. baltica gpgS and mggA genes in E. coli resulted in a high level production of recombinant proteins which were virtually pure. MggB was purified about 43-fold, to a specific activity of 0.013 ± 0.1 μmol/min.mg. This preparation had a major protein band with a molecular mass of 36 kDa corresponding to MggB and two minor bands with lower molecular masses. Attempts to further improve the purity of the protein led to the loss of activity.
The 478-amino acid sequence assigned as GpgS in the R. baltica genome (RB1005) had 80 additional amino acids at the beginning of the sequence without homology with known GpgSs. In fact, no activity was detected for this recombinant protein. We identified a start codon (ATG) 240 bp downstream the original one and new primers were designed in order to amplify the gene excluding this start region. The corresponding recombinant 398-amino acid enzyme had the expected activity.
Activity assays carried out with recombinant -GpgS, -MggA and -MggB showed GPG synthesis by GpgS, MGPG synthesis by MggA and MGPG dephosphorylation by MggB. These activities were also confirmed in assays carried out with R. baltica cell extracts. Conversely, no activity was detected for the MggS, either in R. baltica cell extracts or by the recombinant enzyme, in experiments including the reactions previously described for the MggS from Ptg. mobilis6. The recombinant SPase yielded the expected activity since it catalyzed the conversion of sucrose to fructose and glucose-1-phosphate. Additionally, we tested the SPase for the ability to synthesize glucosylglycerate (GG), resembling the activity of the SPase from Leuconostoc mesenteroides10. However, this activity was not detected, either in R. baltica cell extracts or by the recombinant enzyme.
Catalytic Properties of Glucosyl-3-phosphoglycerate Synthase (GpgS)
The GpgS gene contains 1197 bp, coding for a polypeptide with 398-amino acids, a calculated molecular mass of 44.99 kDa and an isoelectric point of 5.43. Analysis by size-exclusion chromatography showed that the recombinant GpgS behaves as a pentamer with a molecular mass of around 218.6 ± 4.09 kDa, unlike the related GpgS from Per. marina and the homo-functional, but sequence unrelated, GpgSs from mycobacteria and Ptg. mobilis, that formed dimers in solution6,10,12.
R. baltica GpgS was rather non-specific for glucosyl donors using several glucose diphosphate nucleosides, with ADP-glucose being by far the preferred substrate (1.1 ± 0.1 μmol/min.mg specific activity), followed by UDP-glucose (0.17 ± 0.30 μmol/min.mg specific activity) and GDP-glucose (0.03 ± 0.01 μmol/min.mg specific activity). Among the substrates examined only 3-PGA could be used as glucosyl acceptor leading to the formation of GPG.
The identification of the reaction product of GpgS as GPG was achieved by NMR spectroscopy. The proton spectrum of the product displayed an anomeric signal at 4.91 ppm with a proton coupling constant of 3.7 Hz, which proves the α configuration of GPG and places this enzyme in the configuration retaining glycosyl tranferase class. The COSY and TOCSY spectra allowed complete assignment of the proton frequencies in the structure of the glucosyl and glyceryl moieties. A Nuclear Overhauser effect spectrum allowed us to confirm the glycosidic link between position one of the glucose and position two of the glycerate moiety, while the 31P-1H HSQC spectrum enabled us to unequivocally locate the phosphate group at position three of the glycerate, with a phosphorus chemical shift of 2.49 ppm.
GpgS had maximum activity at 60°C, becoming undetectable below 15 and above 90°C. At R. baltica optimal growth temperature the recombinant GpgS retained 30% of the maximal activity (Fig. 1a). The activity of the enzyme at 60 and 25°C was maximal around pH 7.5 (Fig. 1b). The half-lives determined at 60 and 25°C were 2.17 and 57.8 hours, respectively. Moreover, the recombinant GpgS was not strictly dependent on divalent cations, contrary to all other characterized GpgSs6,9,10,12,13, since in the presence of 2.5 mM EDTA the enzyme retained 19% of its maximal activity. The maximum activity was obtained with 1.0 mM Mg2+, at both 60 and 25°C. Nevertheless, the presence of Co2+ and Mn2+ also stimulated its activity (10 mM), although higher Mg2+ concentrations (10–50 mM) had an inhibitory effect (Supplementary Fig. S2).
Temperature and pH dependence of the recombinant GpgS, MggA and MggB from R. baltica.
Temperature (a) and pH dependence (b) of the recombinant GpgS ( ), recombinant MggA (
), recombinant MggA ( ) and recombinant MggB (
) and recombinant MggB ( ) from R. baltica. pH dependence was calculated at recombinant enzyme optimal temperature (filled lines) and at R. baltica optimal growth temperature (dashed lines). Data are the mean values of three independent experiments.
) from R. baltica. pH dependence was calculated at recombinant enzyme optimal temperature (filled lines) and at R. baltica optimal growth temperature (dashed lines). Data are the mean values of three independent experiments.
Kinetic experiments showed that GpgS exhibited Michaelis-Menten kinetics at 60°C, with a Km for ADP-glucose and 3PGA of 9.48 ± 2.59 mM and 4.45 ± 2.11 mM, respectively and a Vmax for ADP-glucose and 3PGA of 2.32 ± 0.09 μmol/min.mg protein and 0.93 ± 0.08 μmol/min.mg protein, respectively. A comparison between the kinetic parameters of R. baltica GpgS and other isofunctional GpgSs determined at each organism's optimum growth temperature are summarized in Table 1.
Catalytic Properties of Mannosylglucosyl-3-phosphoglycerate Synthase (MggA)
The MggA gene contains 1593 bp, coding a polypeptide with 530-amino acids, a calculated molecular mass of 59.7 kDa and an isoelectric point of 5.5. Gel filtration experiments indicated that the recombinant MggA protein behaves as a dimeric protein with a molecular mass of around 121.9 ± 2.04 kDa, unlike the related MggA from Ptg. mobilis that formed monomers in solution6.
R. baltica MggA was non-specific for the mannosyl-donor, using GDP-mannose (3.1 ± 0.5 μmol/min.mg specific activity), UDP-mannose (1.72 ± 0.2 μmol/min.mg specific activity) and ADP-mannose (1.57 ± 0.4 μmol/min.mg specific activity), as substrates with decreasing affinity. From the tested substrates only GPG was used by MggA as mannosyl-acceptor leading to the formation o MGPG.
The identification of the reaction product of MggA as MGPG was achieved by NMR spectroscopy. A combination of 2D spectra (COSY, TOCSY, 13C HSQC and 13C HMBC) allowed us to assign all proton and carbon signals of the glucosyl, mannosyl and glyceryl moieties (Supplementary Table S2 and Supplementary Fig. S3). Furthermore from the HMBC spectrum it was possible to observe conectivities between position two of the glycerate and position one of the glucose; and between position two of the glucose and position one of the mannose moiety, confirming the 1–2 glycosidic bonds, as expected and from the 31P-1H HSQC spectrum we were able to locate the phosphate group at position three of the glycerate, thus fully establishing the structure of the product as mannosyl-glucosyl-phopho-glycerate. The anomeric signals in the proton spectrum at 5.20 and 5.13 ppm presented proton coupling constant of 3.27 and 1.42 Hz, respectively, for the glucose and mannose. Subsequent dephosphorylation (see below) of this product rendered MGG, which is in the α,α configuration of the hexoses, meaning that this enzyme is also a configuration retaining sugar tranferase.
At 25°C the activity of MggA was 40% of the maximal activity reached at 40°C, while at 90°C the enzyme still retained 18% of the total activity (Fig. 1a). Within the pH range examined (5.0 to 10.0), the activity of the enzyme, at both 25 and 40°C, was maximal near pH 7.5 (Fig. 1b). Additionally, the enzyme had a half-life of 52.8 min and 38.5 hours at 40 and 25°C, respectively. The activity of MggA was not strictly dependent on divalent cations. In fact, after incubation with 5.0 mM of EDTA, the enzyme kept 47.5% of its maximum activity. The maximum activation was obtained with 0.1 mM Mn2+, at 25 and 40°C. Additionally, the presence of 1 mM of Co2+ and 10 mM of Ca2+ highly stimulated the MggA activity (Supplementary Fig. S2).
MggA exhibited Michaelis-Menten kinetics at 40°C, with Km values for GDP-mannose and GPG of 0.28 ± 0.04 mM and 0.41 ± 0.07 mM, respectively and similar Vmax for GDP-mannose and GPG of 3.87 ± 0.07 μmol/min.mg protein and 3.46 ± 0.12 μmol/min.mg, respectively. A comparison of kinetic parameters between MggA from R. baltica and from the Ptg. mobilis homologue determined at each organism's optimum growth temperature is summarized in Table 1.
Properties of the partially purified recombinant Mannosylglucosyl-3-phosphoglycerate Phosphatase (MggB)
The MggB gene from R. baltica contains 975 bp coding for a polypeptide with 324 amino acids with a calculated molecular mass of 35.9 kDa and an isoelectric point of 5.7.
MggB activity was strictly dependent on the tested substrate and only MGPG was dephosphorylated to MGG (0.013 ± 0.1 μmol/min.mg specific activity). The identification of MggB reaction product as MGG was achieved by NMR spectroscopy by comparison of the spectral features with the ones from MGG spectra extracted from Ptg. miotherma5. The α,α configuration of the hexoses was verified by the measurement of the coupling constants between the anomeric carbon and the directly bound proton (1JC,H ) of 171.4 and 170.3 Hz, for the mannosyl and glucosyl moieties, respectively.
MggB maximal activity was reached at 40°C and only 28% was retained at 60°C. At R. baltica optimal growth temperature, MggB retained 64% of the maximal activity. Furthermore, no activity was detected below 15°C (Fig. 1a). The pH range for activity at 40°C varied between 4.0 and 9.0 with the optimum pH around 6.0 (Fig. 1b). An identical profile was observed at 25°C. The half-lives determined at 40 and 25°C were 5.3 and 9.6 hours, respectively. MggB lack of activity after the addition of 2.0 mM EDTA indicated that it was strictly dependent on divalent cations in the following order of efficiency: Mg2+ > Ni2+ > Co2+. Maximum activity was reached in the presence of 10 mM of Mg2+ followed by 10 mM of Ni2+; other cations tested, such as K+ and Na+, also provided a good activation of the enzyme (Supplementary Fig. S2). Several examples of the stimulatory effect by divalent ions as well as by monovalent ions were reported, namely in the M. burtonii glucosyl-3-phosphoglycerate phosphatase (GpgP), in Pyrococcus horikoshii mannosyl-3-phosphoglycerate synthase (MpgP) and more recently in cyanobacterial GpgSs9,13,14. It is well documented that K+ is central in the signaling cascade between salt stress sensing and osmoadaptation in prokaryotes in general and archaea in particular15. The physiological significance of the observation that potassium stimulates the dephosphorylation of MGPG argues in favor that the formation of MGG is osmotically regulated.
Kinetics experiments showed that MggB exhibited Michaelis-Menten kinetics at 25 and 40°C. Comparing the Km values at both temperatures for MGPG, 0.22 ± 0.05 mM at 25°C and 6.16 ± 0.08 mM at 40°C, a 28-fold increase was observed with the temperature upshift, evidencing that MggB is functionally adapted to the organism's environment.
Homologues of GpgS, MggA and MggB
The R. baltica GpgS, MggA and MggB proteins were used as seeds for performing BLAST searches in the NCBI and JGI protein databases. Maximum likelihood (ML) phylogenetic trees were obtained for each locus separately with PhyML 3.016.
Genes responsible for GG synthesis were identified in the archaeon M. burtonii9, in two bacteria of the aquificales6,10, in three actinobacteria12,17 and in two cyanobacteria13. The alignment of selected GpgS sequences revealed high sequence conservation despite the large number of sequences found. GpgS homologues were also detected in the genomes from two other members of the Planctomycetes, namely in R. baltica WH47 (GenBank accession number EGF24278) and in Planctomyces maris (NCBI Reference Sequence ZP_01854245). The dichotomy between archaea and bacteria was not observed in the ML tree constructed based on the alignment of known GpgSs and selected homologues since four major clusters with very high bootstrap values were predicted (Supplementary Fig. S4). One cluster includes organisms that have the genes for the second step of MGG synthesis, namely MggA; another cluster comprises GpgS sequences present in all the genomes of the Halobacteriaceae; another is composed primarily of actinobacterial GpgSs, while the remaining GpgS sequences form a different cluster that includes many unrelated bacteria and methanogens (Supplementary Fig. S4). Surprisingly, the homology between the GpgS from R. baltica (rb1005) and those from Ptg. mobilis (NCBI Reference Sequence YP_001568184) and Ptg. miotherma (GenBank accession number ACZ57772), the only known organisms to accumulate MGG5,6, was rather low suggesting a distinct origin. Indeed, these GpgS clustered in distinct groups evidencing their evolutionary differences (Supplementary Fig. S4).
The BLAST searches showed that the gene encoding MggA in R. baltica (gene rb5546) was extremely rare with only twelve low identity homologues. The only functionally characterized MggA so far belongs to the slightly thermophilic member of the deep-branching bacteria of the order Thermotogales, Ptg. mobilis6 and shares 27% amino acid identity with the MggA from R. baltica. The amino acid identity of R. baltica MggA varied between 32% with homologues from the deltaproteobacterium Desulfobulbus propionicus and from the verrucomicrobium Coraliomargarita akajimensis and between 28 and 26% with those from the Spirochaetaceae family (Supplementary Fig. S5). Only one MggA homologue was detected in the genomes from Planctomycetes, namely from R. baltica WH47, with high sequence homology (96%).
Using MggA homologues, an ML tree was constructed displaying two major and distinct groups, each comprising one of the two fully characterized MggAs, one in this study and one from Ptg. mobilis6 (Supplementary Fig. S5). The majorities of the homologues from other organisms are, however, not known to accumulate MGG, nor are the majority of these proteins known to catalyze the synthesis of MGPG. Nevertheless, the high homology between these putative MggAs proteins indicates a common function. Interestingly, all these twelve organisms also possess putative GpgSs, reinforcing the existence of a functional pathway for MGG biosynthesis.
From the genomes, homologues for the MggB (gene rb12347) from R. baltica were also extremely unusual, with only twelve homologues (Supplementary Fig. S6). From these, only nine microorganisms also had homologues for the two other proteins involved in R. baltica MGG synthesis (Supplementary Table S3). The highest homology found was between this enzyme and members of the genus Spirochaeta (32 to 39% amino acid identity). MggB homologues were also detected in the genomes of other members of the Planctomycetes, but interestingly some organisms lacked GpgS homologues, namely Pla. limnophilus and Gemmata obscuriglobus, or lacked MggA homologues, namely Pla. maris. An ML tree constructed based on R. baltica MggB and homologues shows clearly that the proteins from the two strains of R. baltica, SH 1 and WH47, were clustered separately (Supplementary Fig. S6). This observation supports the hypothesis of additional functions for MggB homologues, since R. baltica, the only organism known to accumulate MGG, clusters in a phylogenetic distinct group when compared with the other MggB-homologues containing organisms (Supplementary Fig. S6).
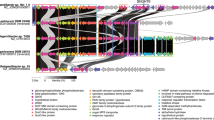
Different genetic organizations of putative gpgS, mggA, and mggB were found in the nine bacteria comprising the three genes (Supplementary Table S3), including isolated genes, consecutive mggA and mggB genes with isolated gpgS, or consecutive gpgS, mggA, and mggB genes with the same orientation in an operon-like organization (Fig. 2). Consecutive mggA and mggB genes with isolated gpgS were found in Candidatus “Desulfococcus oleovorans Hxd3” and in Spirochaeta caldaria DSM 7334, while consecutive gpgS, mggA, and mggB genes with the same orientation in an operon-like organization were only identified in Coraliomargarita akajimensis DSM 45221. In R. baltica, the three genes were also isolated in the chromosome, but with distinct orientations (Fig. 2).
Comparison of the genomic organization and flanking regions of gpgS, mggA and mggB genes from R. baltica and homologues.
Schematic comparison of the genomic organization and flanking regions of gpgS, mggA and mggB genes from R. baltica and homologues described in Supplementary Table S3. Arrows represent identified or putative gpgS (yellow arrows); identified or putative mggA (light blue arrows); identified or putative mggB (dark blue arrows); unspecified genes (white arrows) and their directions.
Phylogeny of MGG synthesizing genes on the basis of concatenated genes analysis
A comparative analysis between the phylogeny obtained from the concatenated alignment of GpgS, MggA and MggB proteins and the corresponding 16S rRNA genes was performed to unravel their evolutionary history (Fig. 3). An important incongruence between the phylogenies of the 16S rRNA gene and the proteins involved in MGG synthesis was identified. The phylogenetic analysis of 16S rRNA genes grouped the organisms into two major clusters, one comprising all members of the Spirochaetaceae family, while the other clustered the remaining microorganisms. On the other hand, the phylogeny of the GpgS/MggA/MggB amino acids clustered the two R. baltica strains into a discrete cluster, while the Candidatus “Desulfococcus oleovorans Hxd3” and C. akajimensis DSM 45221 were grouped with the members of the family Spirochaetaceae. Additionally, a phylogenetic comparison between the previously obtained concatenated alignment of gpgS/mggA/mggB clustered genes and the corresponding deduced amino acid sequences was also performed (results not shown). The clusters inferred from the nucleotide-based subgroups were consistent with the previously obtained from the deduced amino acid sequences (Fig. 3). These findings indicate that most nucleotide polymorphisms resulted in amino acid changes.
Comparison between topologies of the maximum likelihood trees showing the phylogenetic relationships of the 16S rRNA gene sequences, based on the analysis of organism bearing MGG synthesizing enzymes, with the most closely related reference sequences of the LTP database 111 (a) and (b) concatenated GpgS/MggA/MggB.
The two major clusters identified by the 16S rRNAs analysis are highlighted. Incongruence's between the topology of the phylogenetic trees are indicated with dotted lines.
Discussion
Two biosynthetic pathways for MGG have been identified in Ptg. mobilis6 and the enzymes provided us with the information to search for homologues in the R. baltica genome. The genetic content and organization of the genes involved in MGG synthesis is quite different in R. baltica and Ptg. mobilis, envisioning the existence of alternative pathways for MGG synthesis. In R. baltica, contrary to what was described for Ptg. mobilis, the glucosyl-3-phosphoglycerate synthase (gpgS) and mannosylglucosyl-3-phosphoglycerate synthase (mggA) genes were not sequential. Moreover, homologues for the glucosylglycerate synthase (GgS) and for the mannosylglucosyl-3-phosphoglycerate phosphatase (MggB), involved in MGG synthesis in Ptg. mobilis, were not found in the R. baltica genome. Additionally, a mannosylglucosylglycerate synthase (mggS) homologue was identified in R. baltica in an operon-like structure along with a putative hydrolase and a sucrose phosphorylase (spasE), absent in Ptg. mobilis.
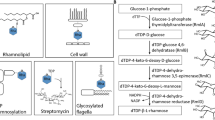
We propose a three-step pathway for the biosynthesis of MGG in R. baltica based on the activity of relevant recombinant enzymes involving the formation of phosphorylated intermediates (Fig. 4). This pathway comprises a GpgS that catalyzes the formation of glucosyl-3-phosphoglycerate (GPG) from ADP-glucose and D-3-phosphoglycerate, which is in turn converted to the phosphorylated precursor of MGG, mannosylglucosyl-3-phosphoglycerate (MGPG), by MggA, closely related to the homologue from Ptg. mobilis6. MGPG is subsequently dephosphorylated to MGG by an uncommon and highly specific mannosylglucosyl-3-phosphoglycerate phosphatase (MggB), which has no homology with the previously described MggB from Ptg. mobilis6.
Proposed pathways for the synthesis of MGG in R. baltica and genomic organization of the involved genes.
Functionally characterized enzymes are in yellow, light and dark blue colors. Dashed lines indicate putative functions. GpgS, glucosyl-3-phosphoglycerate synthase; MggA, mannosylglucosyl-3-phosphoglycerate synthase; MggB, mannosylglucosyl-3-phosphoglycerate phosphatase; Spase, sucrose phosphorylase and MggS, mannosylglucosylglycerate synthase. The MGG structural representation has been adapted from6.
We also hypothesize the existence of an original and alternative nonphosphorylating pathway for the synthesis of MGG in R. baltica based on the existence of homologues for MggS and SPase arranged in the same operon-like structure together with the functionally characterized MggB (Fig. 4). The existence of this putative pathway is supported by the ability of the SPase homologue from L. mesenteroides to successfully synthesize GG in vitro from sucrose and glycerate11. In fact, it was recently hypothesized that this SPase, that is commonly associated with GG synthesizing genes, could represent a third potential pathway for GG synthesis4. The second step of this putative pathway is supported by the ability of the MggS homologue from Thermotoga maritima to synthesize MGG directly from GG and GDP-mannose6. However, we could not functionally express the putative MggS, even after the construction of synthetic genes with optimized sequence for expression in E. coli. Since planctomyces are compartimentalized bacteria the lack of MggS expression could be related with a sub-cellular localization signal present in the protein that could not be recognized by E. coli, so further studies are needed to unravel the real role of this enzyme.
The theoretical existence of two pathways for the synthesis of MGG in R. baltica highlights the importance of MGG as a compatible solute in this microorganism. The same dual pathway mechanism has been described for the synthesis of the related solute GG in Per. marina, for the synthesis of MG in Rhodothermus marinus and more recently, for the synthesis of MGG in Ptg. mobilis6,18,19. In fact, it was demonstrated that the two alternative pathways for the synthesis of MG in Rdt. marinus were differently regulated at the level of expression to be consistent with specific roles in the adaptation to two different types of stress, namely, thermal and osmotic stress20. The reasons underlying the existence of two distinct pathways for the synthesis of MGG in R. baltica remains unclear, but it is conceivable that these systems could be differentially regulated.
R. baltica GpgS has several distinct biochemical and kinetic properties when compared with those previously characterized6,9,10,12,13. The GpgS from R. baltica, like the homologues from Ptg. mobilis, Per. marina, Myc. smegmatis and Myc. bovis, was non-specific for glucosyl donors, using several glucose diphosphate nucleosides, with ADP-glucose being by far the preferred substrate. These results are in contrast to the total dependence of the GpgSs from M. burtonii and from cyanobacteria for GDP-glucose and ADP-glucose, respectively6,9,10,12,13. This trend was also observed with R. baltica MggA that, unlike the isofunctional MggA from Ptg. mobilis6, was non-specific for the mannosyl-donor, using GDP-mannose, ADP-mannose as well as UDP-mannose. This broad substrate specificity may reflect an absolute requirement of MGG in R. baltica, as observed for other glucosyltranferases involved in trehalose synthesis in mycobacteria and sucrose synthesis in cyanobacteria21. Interestingly, the R. baltica recombinant GpgS was not strictly dependent on divalent cations in contrast to their counterparts6,9,10,12,13. Since the synthesis of MGG and glucosylglycerate (GG) have a common step catalyzed by the GpgS, these pathways share, to some extent, the same evolutionary origin. The increasing numbers of genomes confirm that, unlike initially suspected, the biosynthesis of GG may be a widely disseminated property since the gene responsible for GG synthesis - gpgS, is widespread throughout most lineages of bacteria. Indeed, gpgS sequences seem to have been preferred by nature over the mggA sequences, since this second group displays a significantly lower occurrence. The presence of both genes in members of the deepest lineage of Bacteria4 strongly supports an ancient origin. The evolution of GG and MGG accumulation appears to be the combined result of mggA gene loss across some branches of the Tree of Life, with conservation of this characteristic in some scattered groups. Hence, the wider distribution of the genes responsible for GG synthesis suggests that this characteristic has been positively selected over MGG in the course of evolution. Indeed, this gene has been identified in halophiles and halotolerant, alkaliphiles, psycrophiles, thermophiles and radio-resistant microorganisms (Supplementary Fig. S4) and new discoveries can be predicted concerning the role of GG in the adaptation of prokaryotes to several environmental challenges.
MggAs from R. baltica and Ptg. mobilis also have several distinct biochemical and kinetic properties. The maximal activities of R. baltica MggA occurred around 40°C (Fig. 1a), representing a thermal shift of 50°C over that from Ptg. mobilis, as expected for a mesophilic organism. Moreover, the pH dependences and the substrate specificities were also quite distinct for both homologues. These differences are supported by the low homology between these two enzymes. MggA from Ptg. mobilis was initially classified as belonging to the GT1 family of inverting glycosyltransferases6, but a more detailed analysis annotated MggA as an α-mannosyltransferase of the GT4 retaining family22,23. Moreover, both MggA and MggS are classic Leloir glycosyltransferases, not synthases.
The MggB from R. baltica dephosphorylated only MGPG to MGG within the substrates examined. Interestingly, this enzyme has no significant sequence homology with other known phosphatases and this divergence may explain the very high specificity of this enzyme for MGPG. MggB has been assigned as a member of the Haloacid dehalogenase-like hydrolase superfamily (HAD-like hydrolase) and residues that compose the conserved active site and motif II (conserved serine) of HAD-like hydrolase24 were mapped in MggB. No homology was detected between the R. baltica and Ptg. mobilis MggB, a previously tentative description for a mannosylglucosyl-3-phosphoglycerate phosphatase putatively responsible for the dephosphorylation of MGPG to MGG in Ptg. mobilis6. MggB homologues were detected in the genomes of other members of the Planctomycetes, but interestingly some organisms lacked GpgS homologues, namely Pla. limnophilus and G. obscuriglobus, or lacked MggA homologues, namely Pla. maris. This observation suggests additional functions for this enzyme depending on the analyzed organism that were not identified for the MggB from R. baltica. Reinforcing this hypothesis is the very low homology found between R. baltica MggB and homologues from the above mentioned planctomycetes.
The phylogenetic distribution of the enzymes and homologues implicated in the synthesis of MGG reveals no obvious correlation between the accumulation of this compatible solute and thermo- or osmo-adaptation. The habitats where these putatively accumulating MGG organisms have been isolated range from sea water, fresh water to termite guts with optimal growth temperatures ranging from 20°C to 66°C (Supplementary Table S3). Interestingly, the α-mannopyranosyl-(1,2)-α-glucopyranose unit found in MGG is also present in the polar head groups of some glycolipids found in extremely halophilic archaea24. Moreover, MGG is structurally similar to the diglucosylglycerate found on the reducing end of the mycobacterial methylglucose lipopolysaccharides (MGLP), precursors of which are also GDP-mannose and GPG. This evidence suggests a more complex role for this compatible solute, as was demonstrated for the related solute GG. This aforementioned multi-tasked molecule acts as a contingency compatible solute under combined salt stress and nitrogen-deficient conditions or as a precursor for more elaborated macromolecules, namely as intermediate to glycolipids or polysaccharides4.
In summary, our results determined that the described pathway is committed to MGG synthesis in R. baltica. Knowledge of the physiological role of MGG in R. baltica will provide a more detailed understanding towards elucidation of the salt stress adaptation mechanisms in this ancient lineage of organisms. Characterization of this pathway involved in the synthesis of MGG in R. baltica is an essential step towards this goal.
Methods
Identification and cloning and functional overexpression of gpgS, mggA, mggB, spasE and putative mggS
R. baltica type strain (DSM 10527) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany and cultured at 25°C in modified M13a medium containing glucose (1.8 g/L) and ammonium sulfate (0.32 g/L). To identify the genes for mannosylglucosylglycerate (MGG) biosynthesis we used the amino acid sequences of glucosyl-3-phosphoglycerate synthase (GpgS) from Ptg. mobilis, from Persephonella marina and from Methanococcoides burtonii (Accession nos. YP_001568184, ABX75857 and ABE51702, respectively); of mannosylglucosyl-3-phosphoglycerate synthase (MggA), mannosylglucosylglycerate synthase (MggS) and of the putative mannosylglucosyl-3-phosphoglycerate phosphatase (MggB) from Ptg. mobilis (Accession nos. YP_001568185, YP_001567745 and YP_001567658, respectively) to carry out BLATP with the scoring matrix BLOSUM62 in the R. baltica genome database (KEGG Genome Database [http://www.genome.jp/kegg/]).
The gpgS, mggA and mggS homologues and the putative hydrolase (mggB) and sucrose phosphorylase (spasE) were amplified using the primers described in Supplementary Table S1. PCR amplifications were performed as previously described26, cloned into the corresponding sites of the expression vector pET30a (Novagen) and transformed into E. coli BL21 using standard molecular biology procedures27. Several other approaches were used to facilitate the purification of the recombinant enzymes but without success. Namely, all genes were cloned without the stop codons into a His-tag containing plasmid - pET30A (Novagen), into pTRC99A (Novagen) and into pTRC99A-GST (12) and pGEX-GST, these last two are gluthatione-S-transferase (GST) containing plasmids. The unsuccessful attempt to express these genes could be due to the presence of numerous codons rarely used by E. coli. Therefore, all genes were synthesized for optimum codon usage for expression in E. coli (GenScript, USA) and the previously cloning protocol was applied once more, but without success. Constructs were sequenced by Macrogen Corporation (Netherlands).
Recombinant E. coli was grown to mid-exponential phase of growth (OD600 = 0.8) in LB medium at 37°C, induced with 0.5 mM IPTG and grown for further 16 h at 25°C, harvested, disrupted, centrifuged to remove debris and protein content was determined28. Kanamycin was added to a final concentration of 30 μg/ml.
Synthesis, analysis, purification and quantification of MGPG, GPG, GG and MGG
Glucosyl-3-phosphoglycerate (GPG) and mannosylglucosyl-3-phosphoglycerate (MGPG) were synthesized overnight using the pure recombinant GpgS from Mycobacterium smegmatis29 and the purified recombinant MggA from Ptg. mobilis6, respectively, under optimal enzyme conditions. GPG and MGPG were purified as previously described8. Glucosylglycerate (GG) was synthesized from GPG, as previously described9. Mannosylglucosylglycerate (MGG) was synthesized from the dephosphorylation of MGPG in a reaction mixture containing MGPG, 2 U of alkaline phosphatase (Promega) for 15 min at 37°C. The GG and MGG synthesis was analyzed by thin layer chromatography (TLC) with a solvent system composed of chloroform, methanol, acetic acid and water (30:15:8:4, v/v). GPG was separated from residual UDP-glucose and MGPG from residual GDP-mannose and GPG by TLC with a solvent system composed of n-propanol, ethyl acetate, water and a 25% ammonium solution (50:10:30:10). Mannose, glucose, GDP, GDP-mannose, UDP-glucose (all from Sigma-Genosys), GPG, MGPG, GG and MGG standards were used for comparative purposes.
Preparation of R. baltica cell extracts and enzyme assays
R. baltica biomass was obtained from a five liter culture under the conditions described above. Cells were harvested during the late exponential phase of growth (9,000 × g, 5 min, 4°C), suspended in 20 mM Tris–HCl (pH 7.6) and disrupted twice with a French-press cell. The supernatant was dialyzed overnight against 20 mM Tris–HCl (pH 7.0).
The GpgS, MggA, MggB and MggS activities in cell extracts were assessed overnight in 20 mM Tris-HCl (pH 7.5) with 10 mM MgCl2. Substrates, 5 mM (each), were D-3-phosphoglycerate (3-PGA) and UDP-glucose for GpgS, GPG and GDP-mannose for MggA, MGPG for MggB and GG and GDP-mannose for MggS. The hydrolytic ability of the putative MggS was determined as previously described6. The SPase activities were tested overnight in cell extract with 5.0 mM of sucrose in 5.0 mM phosphate buffer (pH 7.0) or with 5.0 mM of morpholineethanesulfonic acid (MES) (pH 7.0). The inverse reaction was also tested by incubating the cell extract with 10 mM of fructose plus 10 mM of glucose-1-phosphate in 25 mM MES (pH 7.0). The described in vitro synthesis of GG by the SPase from Leuconostoc mesenteroides was also investigated, as previously described11.
Purification of recombinant GpgS, MggA and MggB
The same purification protocol was used for the recombinant GpgS and MggA, with minor variations. The recombinant containing supernants were loaded onto a DEAE Sepharose column equilibrated with 20 mM Tris-HCl (pH 7.5 and pH 7.0 for the recombinant GpgS and MggA, respectively) and eluted by a linear gradient of 0.0 to 1.0 M NaCl. Active fractions were pooled, concentrated by centrifugation in 30 kDa cutoff centricons, loaded onto a Q-Sepharose column equilibrated with 20 mM BTP (pH 8.0) and eluted in a linear NaCl gradient (0.0 to 0.5 M), in the same buffer. Desalted samples were loaded in a Q-Resource column in 20 mM BTP (pH 8.0) and eluted by a linear gradient of 0.0 to 0.3 M NaCl. For GpgS, two consecutive columns were used under the same conditions. Active fractions were concentrated and equilibrated in 50 mM Tris-HCl with 200 mM NaCl (pH 7.5) by ultrafiltration, as previously described and loaded onto two consecutive Superdex 200 columns.
Recombinant MggB was partially purified by anion-exchange with two sequential Q-Resource columns equilibrated with 20 mM Tris-HCl (pH 7.0) and eluted by a linear gradient of 0.0 to 0.4 M NaCl. The active fractions were pooled, concentrated and equilibrated in 20 mM Tris-HCl (pH 7.0) by ultrafiltration, as described above. In an attempt to get better yields of purification several anion-exchange and gel-filtration chromatographies columns were tested, namely Q-Sepharose, DEAE Sepharose and Superdex-200, as described above, all resulting in a complete loss of MggB activity.
The purity of each of the recombinant GpgS, MggA and MggB was judged by SDS-PAGE (Supplementary Fig. S1).
Characterization of recombinant GpgS
Temperature profile, pH dependence, effect of cations, thermal stability, substrate specificity and molecular mass of recombinant GpgS were measured from the synthesis of GPG, as previously described10. The temperature profile was determined between 25 and 90°C in 25 mM BTP (pH 7.5) with 1.0 mM MgCl2. The effect of pH was determined at 60°C in 25 mM acetate (pH 4.0–5.0), 25 mM BTP (pH 6.5–8.0), 25 mM Tris-HCl buffer (pH 7.0–9.0) and 25 mM CAPSO (pH 9.0–10.0). All pH values were measured at room temperature (25°C); the pH at 60°C was calculated using a conversion factor (ΔpKa/ΔT = 0.0002 for acetate buffer, 0.015 for BTP, 0.03 for Tris-HCl and 0.009 for CAPSO)30,31. All other parameters were determined at 60°C in 25 mM BTP (pH 7.5) with 1.0 mM MgCl2.
The specific activity for each sugar donor used was determined with 2.5 mM concentration of each substrate. The determined parameters for optimal enzyme activity were also tested at 25°C, as described above. The thermal stability was determined in 25 mM BTP (pH 7.5) at 25 and 60°C. At appropriate times, samples were withdrawn and immediately examined for residual activities at 25°C and 60°C, respectively.
The kinetic parameters were determined at 25°C and at 60°C, in reaction mixtures contained ADP-glucose (1.0 mM to 20 mM) plus 3-PGA (10 mM) or ADP-glucose (10 mM) plus 3-PGA (1.0 mM to 15 mM), as previously described10. All experiments were performed in triplicate.
Characterization of recombinant MggA
The assay used for characterization was based on the quantification of NDPs released during the conversion of NDP-mannose and GPG to MGPG. Reactions mixtures containing 25 mM Tris-HCl (pH 7.5) with 0.1 mM MnCl2, 2.0 mM GDP-mannose and 2.0 mM GPG, were initiated by adding exact amounts of MggA and stopped at different times by cooling on ethanol-ice. The amount of NDP released was determined as previously described32. All parameters, except the temperature profile, were determined at 40°C. The temperature profile was evaluated between 10 and 90°C. The pH dependence, effect of cations and molecular mass were examined as previously described6. The specific activity for each sugar donor used was determined as described above, with a 2.5 mM concentration of each substrate. The thermal stability was determined at 25°C and at 40°C. At appropriate times, samples were reserved and immediately examined for residual activities at 25°C and 40°C, respectively. The determined parameters for optimal enzyme activity were also tested at 25°C, as described above.
The kinetic parameters for GDP-mannose and GPG were determined by using 25 mM Tris-HCl (pH 7.5) with 0.1 mM MnCl2 and 3 μg of MggA, at 25°C and 40°C, containing either GDP-mannose (1.0 mM to 12 mM) plus GPG (10 mM) or GDP-mannose (10 mM) plus GPG (1.0 mM to 10 mM). All experiments were performed in triplicate.
Characterization of recombinant MggB
The substrate specificity was determined using MGPG, GPG, MPG, ADP, GDP, UDP, 2-PGA, 3-PGA, UDP-glucose, ADP-glucose, GDP-glucose, GDP-fucose, GDP-mannose, glucose-1-phosphate, glucose-6-phosphate, fructose-6-phosphate, mannose-1-phosphate and mannose-6-phosphate as substrates. Reaction mixtures containing 2.0 mM of each substrate and 10.0 mM MgCl2 in 25 mM Tris (pH 7.0) were incubated overnight at 25°C. The substrate hydrolysis was visualized by TLC, as described above. The assay used for the characterization was based on the release and quantification of inorganic phosphate33 during the hydrolysis of MGPG to MGG. Reactions mixtures containing the appropriate buffer and 5.0 mM of MGPG were initiated by adding exact amounts of MggB and stopped at different times by cooling on ethanol-ice.
The temperature profile was determined between 10 and 70°C with 10 mM MgCl2. All other parameters were evaluated at 40°C. The effect of pH on the activity was determined in 25 mM acetate buffer (pH 4.0–5.0), 25 mM MES (pH 6.0), 25 mM Tris-HCl buffer (pH 7.0–8.0) and 25 mM BTP (pH 7.0–9.0), with 10 mM MgCl2. The cation dependence was examined by incubating the sample with the appropriate substrates with 1.0 mM, 10 mM, 50 mM and 100 mM concentration of Mg2+, Mn2+, Co2+ and 200 mM or 500 mM concentration of K+, Na+, without cations or divalent cations or with 2.0 mM of EDTA. The determined parameters for optimal enzyme activity were also tested at 25°C, as described above. The thermal stability was determined at 40 and 25°C in 25 mM MES (pH 6.0) with 10 mM MgCl2. At appropriate times, samples were reserved and immediately examined for residual activities at 25°C and 40°C, respectively.
The kinetic parameters were determined at 40°C and at 25°C in reaction mixtures containing MGPG (0.1 to 8.0 mM) in 25 mM MES buffer (pH 6.0) with 10 mM MgCl2. All experiments were performed in triplicate.
NMR spectroscopy
GPG, MGPG, GG and MGG were quantified by 1H-nuclear magnetic resonance after freeze-drying and dissolution in 2H2O. All spectra were acquired on a Bruker AVANCE II 500 spectrometer (Bruker, Rheinstetten, Germany) working at a proton operating frequency of 500.43 MHz, equipped with a two channel 5 mm inverse detection probe head with pulse-field gradients along the Z axis. 1H and 13C chemical shifts were referenced to 3 (trimethylsilyl)propanesulfonic acid (sodium salt), while 31P was referenced to external phosphoric acid (85%).
1H-NMR spectra were acquired at 27°C using a 90° flip angle, with water presaturation during a relaxation delay of 2.5 s. For quantification purposes, an extra delay of 60 s was used to allow complete signal relaxation and sodium formate was added as an internal concentration standard. Two dimensional spectra (correlation spectroscopy – COSY; total correlation spectroscopy – TOCSY; nuclear Overhauser effect spectroscopy – NOESY; heteronuclear single quantum coherence spectra – HSQC; and heteronuclear multiple bond correlation – HMBC) were acquired using standard Bruker pulse programs.
GpgS, MggA and MggS sequences
R. baltica GpgS, MggA and MggS sequences (Acc. No. NP_864186, NP_866758 and NP_870458, respectively) were used as seed for BLASTP searches in NCBI database with an expected threshold of 10.
Alignment and phylogenetic reconstruction
A multiple alignment of amino acid sequences was obtained using ClustalΩ35 and manually corrected where necessary. Additionally, for further phylogenetic analyses, GpgS, MggA and MggB proteins were concatenated in a single multiple alignment with BioEdit 7.0.9.036. The MEGA5 package37 was used to derive the multiple alignments of nucleotide and positions of doubtful homology were removed using Gblocks38. Maximum likelihood (ML) phylogenetic analysis were performed for the amino acid concatenated alignment and for each individual locus by PhyML 3.016, using the most appropriate model of amino acid substitution and likelihood scores assessed by ProtTest 2.439. ML phylogenetic analysis was also performed for the nucleotide concatenated alignment using the most appropriate model of nucleotide substitution and likelihood scores assessed by TOPALi V2.540 and by jModeltest41. The best model was determined by using the Akaike Information Criterion (AIC)42.
The phylogenetic inference of the 16S rRNA gene was performed using the ARB software package using the Living Tree Project (LTP) release 111 (http://www.arb-silva.de/projects/living-tree/). The LTP dataset was used for the automatic alignment of the sequences using the SINA aligner followed by a manual curation of misplaced bases. The sequences from Rhodopirellula baltica WH47, Candidatus “Desulfococcus oleovorans Hxd3”, Treponema primitia ZAS-2 and Spirochaeta thermophila DSM 6578 were inserted into the LTPs111 tree using the ARB-parsimony tool implemented in the ARB34 software package and closely related reference sequences were selected. A de novo phylogenetic reconstruction of isolates using almost full length sequences with the reference sequences was calculated using the RAxML algorithm with the GTRGAMMA model.
References
Fuerst, J. A. & Sagulenko, E. Keys to eukaryality: planctomycetes and ancestral evolution of cellular complexity. Front Microbiol 3, 167–178 (2012).
Fuerst, J. A. & Sagulenko, E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9, 403–413 (2011).
da Costa, M. S., Santos, H. & Galinski, E. A. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv Biochem Eng Biotechnol 61, 117–153 (1998).
Empadinhas, N. & da Costa, M. S. Diversity, biological roles and biosynthetic pathways for sugar-glycerate containing compatible solutes in bacteria and archaea. Environ Microbiol 13, 2056–2077 (2010).
Jorge, C., Lamosa, P. & Santos, H. α-D-Mannopyranosyl-(1 —> 2)-α-D-glucopyranosyl-(1 —> 2)- glycerate in the thermophilic bacterium Petrotoga miotherma - structure, cellular content and function. The FEBS J 274, 3120–3127 (2007).
Fernandes, C. et al. Two alternative pathways for the synthesis of the rare compatible solute mannosylglucosylglycerate in Petrotoga mobilis. J Bacteriol 192, 1624–1633 (2010).
d′Avó, A. F., Cunha, S., Mingote, A., Lamosa, P., da Costa, M. S. & Costa, J. A unique pool of compatible solutes from Rhodopirellula baltica a member of the deep-branching phylum Planctomycetes. PLoS ONE 8(6), e68289 (2013).
Schlesner, H. et al. Taxonomic heterogeneity within the Planctomycetales as derived by DNA–DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 54, 1567–1580 (2004).
Costa, J., Empadinhas, N., Lamosa, P., Santos, H. & da Costa, M. S. Characterization of the biosynthetic pathway of glucosylglycerate in the archaeon Methanococcoides burtonii. J Bacteriol 188, 1022–1030 (2006).
Costa, J., Empadinhas, N. & da Costa, M. S. Glucosylglycerate biosynthesis in the deepest lineage of the Bacteria: characterization of the thermophilic proteins GpgS and GpgP from Persephonella marina. J Bacteriol 189, 1648–1654 (2007).
Sawangwan, T., Goedl, C. & Nidetzky, B. Single-step enzymatic synthesis of (R)-2-O-α-D-glucopyranosyl glycerate, a compatible solute from micro-organisms that functions as a protein stabiliser. Org Biomol Chem 7, 4267–4270 (2009).
Empadinhas, N., Albuquerque, L., Mendes, V., Macedo-Ribeiro, S. & da Costa, M. S. Identification of the mycobacterial glucosyl-3-phosphoglycerate synthase. FEMS Microbiol Lett 280, 195–202 (2008).
Klähn, S., Steglich, C., Hess, W. R. & Hagemann, M. Glucosylglycerate: a secondary compatible solute common to marine cyanobacteria from nitrogen-poor environments. Environ Microbiol 12, 83–94 (2010).
Empadinhas, N., Marugg, J. D., Borges, N., Santos, H. & da Costa, M. S. Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. Biochemical and genetic characterization of key enzymes. J Biol Chem 276, 43580–43588 (2001).
Roberts, M. F. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems 1: 5 (2005).
Guindon, S. & Gascuel, O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704 (2003).
Empadinhas, N. et al. Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity. Mol Microbiol 79, 76–93 (2011).
Fernandes, C., Empadinhas, N. & da Costa, M. S. Single-step pathway for synthesis of glucosylglycerate in Persephonella marina. J Bacteriol 189, 4014–4019 (2007).
Martins, L. O. et al. Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus: biochemical and genetic characterization of a mannosylglycerate synthase. J Biol Chem 274, 35407–35414 (1999).
Borges, N., Marugg, J. D., Empadinhas, N., da Costa, M. S. & Santos, H. Specialized roles of the two pathways for the synthesis of mannosylglycerate in osmoadaptation and thermoadaptation of Rhodothermus marinus. J Biol Chem 279, 9892–9898 (2004).
Elbein, A. D., Pan, Y. T., Pastuszak, I. & Carroll, D. New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–27R (2003).
Coutinho, P. M., Deleury, E., Davies, G. J. & Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328, 307–317 (2003).
Campbell, J. A., Davies, G. J., Bulone, V. & Henrissat, B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326, 929–939 (1997).
Koonin, E. V. & Tatusov, R. L. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J Mol Biol 244, 125–132 (1994).
Koga, Y. & Morii, H. Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci Biotechnol Biochem 69, 2019–2034 (2005).
Costa, J., D'Avó, A. F., da Costa, M. S. & Veríssimo, A. Molecular evolution of key genes for type II secretion in Legionella pneumophila. Environ Microbiol 14, 2017–2033 (2012).
Green, M. R. & Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed. (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.: 2012).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248–254 (1976).
Mendes, V., Maranha, A., Alarico, S., da Costa, M. S. & Empadinhas, N. Mycobacterium tuberculosis Rv2419c, the missing glucosyl-3-phosphoglycerate phosphatase for the second step in methylglucose lipopolysaccharide biosynthesis. Scientific Reports 1, 10.1038/srep00177 (2011).
Good, N. et al. Hydrogen ion buffers for biological research. Biochemistry 5, 467–477 (1966).
Mana-Capelli, S., Mandal, A. K. & Arguello, J. M. Archaeoglobus fulgidus CopB is a thermophilic Cu2-ATPase. J Biol Chem 278, 40534–40541 (2003).
Qu, Q., Lee, S.-J. & Boos, W. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem 279, 47890–47897 (2004).
Ames, B. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8, 115–118 (1966).
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539 (2011).
Hall, T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41, 95–98 (1999).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17, 540–552 (2000).
Abascal, F., Zardoya, R. & Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005).
Milne, I. et al. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 20, 1806–1807 (2004).
Posada, D. jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253–1256 (2008).
Akaike, H. A new look at the statistical model identification. IEEE Trans Autom Control 19, 716–723 (1974).
Acknowledgements
This work was supported by Fundação para a Ciência e Tecnologia (FCT) Project PTDC/BIA-MIC/105247/2008 and PEst-C/SAU/LA0001/2011. S. Cunha acknowledge scholarship from FCT (SFRH/BD/46212/2008). The National NMR Network (REDE/1517/RMN/2005) was supported by POCI 2010 and FCT. We are especially grateful to Helena Santos and her group (ITQB, Oeiras, Portugal) for valuable collaboration. We wish to thank Luis França (Center for Neuroscience and Cell Biology, University of Coimbra) for advice on the phylogenetic analyses.
Author information
Authors and Affiliations
Contributions
S.C., A.F.d.A., A.M., P.L. and J.C. performed the experiments. S.C., J.C. and M.S.d.C. conceived and designed the experiments and analyzed the data. S.C., P.L., M.S.d.C. and J.C. wrote the paper. M.S.d.C. contributed reagents/materials/analysis tools. All authors reviewed the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cunha, S., d'Avó, A., Mingote, A. et al. Mannosylglucosylglycerate biosynthesis in the deep-branching phylum Planctomycetes: characterization of the uncommon enzymes from Rhodopirellula baltica. Sci Rep 3, 2378 (2013). https://doi.org/10.1038/srep02378
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02378
This article is cited by
-
Impact of Inoculation with Pseudomonas aestus CMAA 1215T on the Non-target Resident Bacterial Community in a Saline Rhizosphere Soil
Current Microbiology (2021)
-
Comparison of the compatible solute pool of two slightly halophilic planctomycetes species, Gimesia maris and Rubinisphaera brasiliensis
Extremophiles (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.