Abstract
To better understand the potential relationship between COVID-19 disease and hologenome microbial community dynamics and functional profiles, we conducted a multivariate taxonomic and functional microbiome comparison of publicly available human bronchoalveolar lavage fluid (BALF) metatranscriptome samples amongst COVID-19 (n = 32), community acquired pneumonia (CAP) (n = 25), and uninfected samples (n = 29). We then performed a stratified analysis based on mortality amongst the COVID-19 cohort with known outcomes of deceased (n = 10) versus survived (n = 15). Our overarching hypothesis was that there are detectable and functionally significant relationships between BALF microbial metatranscriptomes and the severity of COVID-19 disease onset and progression. We observed 34 functionally discriminant gene ontology (GO) terms in COVID-19 disease compared to the CAP and uninfected cohorts, and 21 GO terms functionally discriminant to COVID-19 mortality (q < 0.05). GO terms enriched in the COVID-19 disease cohort included hydrolase activity, and significant GO terms under the parental terms of biological regulation, viral process, and interspecies interaction between organisms. Notable GO terms associated with COVID-19 mortality included nucleobase-containing compound biosynthetic process, organonitrogen compound catabolic process, pyrimidine-containing compound biosynthetic process, and DNA recombination, RNA binding, magnesium and zinc ion binding, oxidoreductase activity, and endopeptidase activity. A Dirichlet multinomial mixtures clustering analysis resulted in a best model fit using three distinct clusters that were significantly associated with COVID-19 disease and mortality. We additionally observed discriminant taxonomic differences associated with COVID-19 disease and mortality in the genus Sphingomonas, belonging to the Sphingomonadacae family, Variovorax, belonging to the Comamonadaceae family, and in the class Bacteroidia, belonging to the order Bacteroidales. To our knowledge, this is the first study to evaluate significant differences in taxonomic and functional signatures between BALF metatranscriptomes from COVID-19, CAP, and uninfected cohorts, as well as associating these taxa and microbial gene functions with COVID-19 mortality. Collectively, while this data does not speak to causality nor directionality of the association, it does demonstrate a significant relationship between the human microbiome and COVID-19. The results from this study have rendered testable hypotheses that warrant further investigation to better understand the causality and directionality of host–microbiome–pathogen interactions.
Similar content being viewed by others
Introduction
Metatranscriptomes from tissues and biologic samples arising from hosts with varying disease severity and outcomes represent a rich source of information to evaluate the role of the microbiome in onset and progression. For respiratory viruses like SARS-CoV-2, bronchoalveolar lavage fluid (BALF) is a valuable sample type collected to investigate the biology of lower respiratory tract infections. Unfortunately, this sample type is more challenging to obtain for research studies that require large numbers of matching cases and controls, especially compared to the more easily accessible sample types like nasopharyngeal swabs. In general, BALF samples arise from patients that either have a clinical indication for them to be obtained or from healthy controls that have consented for the procedure. Early in the SARS-CoV-2 outbreak, scientists published metatranscriptome sequences from BALF of patients with COVID-19 disease and made the data available in the public domain (Suppl. Table 1); however, limitations in the sample numbers and lack of uniformity in study designs across different laboratories prevented a robust statistical analysis from taking place. In this paper, we computationally evaluate microbial insights drawn from these valuable BALF samples, despite the experimental study design limitations. In contrast to other studies that focus on characteristics of the human host response or SARS-CoV-2 lineages and viral variants, our analysis specifically evaluated the microbial taxonomic and functional profiles of the BALF metatranscriptomes. The role of the human microbiome in SARS-CoV-2 infection is poorly understood, but it remains important to study, since it could be a significant contributor to the observed variations in COVID-19 disease severity and resiliency between patients.
Among other risk factors, it is possible that the lower respiratory tract microbiome plays a role in COVID-19 disease severity. A previous 16S rRNA gene study found that COVID-19 patient endotracheal aspirates had lower microbial diversity compared to uninfected individuals, but these differences were not found to have a significant impact on fatality outcomes1. The original Shen et al. study2 performed a microbial taxonomic analysis of sequenced BALF metatranscriptomes without evaluating functional profiles or considering COVID-19 disease severity in the microbial analysis. Haiminen et al.3 reanalyzed BALF metatranscriptome sequences from the Shen et al. study2 and identified differences in expressed metabolic pathways in COVID-19 samples compared to the uninfected and community acquired pneumonia (CAP) cohorts; however, functional profile differences were not analyzed based on COVID-19 clinical severity. Yang et al.4 analyzed previously published BALF metatranscriptome datasets from multiple independent studies2,5,6,7,8,9,10 and performed a comparative taxonomic analysis between samples from COVID-19 patients and healthy control groups but did not subdivide cohorts further or perform functional analyses. Other studies have focused solely on the taxonomic analysis of a subset of published BALF metatranscriptomes and specific potential co-infections that may be present11,12. To our knowledge, this study is the first to evaluate significant differences in taxonomic and functional signatures between BALF metatranscriptomes from COVID-19, CAP, and uninfected cohorts, as well as COVID-19 morbidity and mortality.
To better understand the potential relationship between COVID-19 morbidity and mortality and the human-microbiome, we conducted an analysis using human BALF metatranscriptome samples sourced from eight publications and nine corresponding public data repositories (Suppl. Tables 1 and 2). BALF specimens from individual subjects were grouped into one of three categorical classes: (1) uninfected controls; (2) community acquired pneumonia (CAP) patients; or (3) COVID-19 patients with moderate to severe disease, including death (Table 1). The objectives of the current study were to compare the BALF metatranscriptomes amongst and between each of the three cohort categorical classes or their sub-categories, such as COVID-19 severe disease versus death, and to identify significantly associated taxonomic and functional differences in microbial derived community dynamics. To achieve these objectives, relevant metatranscriptome datasets were compiled from public sources and a rigorous analysis pipeline was implemented to assess (1) the composition of the microbiome taxa in association with respiratory disease and (2) the microbial gene functions significantly perturbed.
Our overarching testable hypothesis was that there is a potentially informative and discernably significant relationship between the BALF microbiome and the severity of COVID-19 disease. We tested this hypothesis with the following aims: (a) identify significantly associated taxonomic differences between each of the three cohort categorical classes (i.e., uninfected, CAP, COVID-19), (b) discern microbiome-derived functional changes attributed to these community dynamics, and (c) assess these taxonomic and functional differences in relation to the COVID-19 disease outcomes of survived vs. deceased.
Methods
Data acquisition and exclusion
Between the beginning of the COVID-19 pandemic and May 2022, we identified five studies with COVID-19 BALF samples and five studies with non-COVID-19 BALF samples (Suppl. Tables 1 and 2). The publicly available Illumina reads were downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) or the China National Center for Bioinformation (CNCB) National Genomics Data Center (NGDC), along with the original publications where the clinical information was obtained for downstream analysis of BALF samples2,5,6,7,8,13,14,15. Sample types of “unknown” and “sick” from Huang et al.14 and Michalovich et al.8 were pruned from subsequent analysis. “Healthy” samples from Michalovich et al.8 and SARS-CoV-2 viral-enriched samples from Shen et al.2 (PRJNA605907) were also pruned from subsequent analysis (Suppl. Tables 1 and 2). Whenever negative controls were present, the R package decontam25 was used to identify and remove potential contaminating organisms. All negative controls (i.e., CRR125995, CRR125996, CRR125997, CRR125998) came from the Shen et al. study, where the negative controls were described in the original publication as either a nuclease-free water sample or saline solutions that passed through the bronchoscope2. After read-filtering and batch-effect sample removal, sample cohorts of n = 29 uninfected samples from 29 subjects, n = 25 CAP samples from 25 subjects, and n = 32 COVID-19 samples from 18 subjects were available for comparison (total n = 86 BALF samples from n = 72 subjects, where a subset of subjects were sampled multiple times). Amongst the COVID-19 cohort at the time of the index study publication, n = 10 samples were from 5 known-deceased subjects, n = 15 samples were from 9 known-survived subjects, and n = 7 from 4 subjects of the total 32 COVID-19 samples in this meta-analysis with unknown / unpublished survival outcomes.
Quality control and data preprocessing
After the raw reads were downloaded from their sources, the quality of the reads was assessed before and after trimming with FastQC16, and quality control (e.g., adapter removal) was performed on the downloaded sequence reads with Trimmomatic17. To control for different sequencing approaches by dataset (e.g., datasets being paired, or single-end reads), all paired-end reads were merged with FLASH18 and concatenated with unmerged reads into one fastq file per sample. Human and PhiX reads were filtered out with a custom Kraken219 database built with solely human and PhiX references (see data and script availability section below), and low-complexity sequences were removed with fastp20.
Taxonomic and functional assignments
After data preprocessing, a taxonomic analysis was subsequently performed with Kraken219 utilizing their standard database. The processed fastq datasets with human and PhiX reads removed were converted to fasta files and analyzed with SeqScreen21 to obtain a list of leaf-node molecular function and biological process Gene Ontology (GO) terms present within each of the samples. The CoV-IRT-Micro conda package (https://github.com/AstrobioMike/CoV-IRT-Micro), along with programs modified from the bit package22, was used to propagate parent GO terms, parse SeqScreen outputs by taxonomic domain, and summarize Kraken2 taxonomic results and SeqScreen-reported protein identifiers. Parent-propagated GO term counts for all domains other than eukaryotes were imported into a working phyloseq23 object alongside collected and curated clinical metadata using R 4.0324. GO term abundances from the remaining subjects’ specimens were compositionally transformed, center log ratio (CLR) normalized, and independently compared by case type (COVID-19 vs. CAP and Uninfected) and survival outcome (COVID-19 only deceased vs. survived) via MaAsLin226 using minimum abundance, prevalence, and significance cutoffs of 0.01, 0.1, and q < 0.05 (Benjamini–Hochberg multiple test correction), respectively27 (Suppl. Tables 3 and 4). Taxonomic differences identified via MaAsLin2 were subsequently compared by case type and survival outcome with heat tree visualizations using log2 median ratio differences using metacoder (v0.34)28. In order to identify and describe any variability for the observed taxonomic and functional features that was distinctive by case type (i.e., COVID-19, CAP, uninfected) or COVID-19 mortality, we employed Dirichlet multinomial mixture (DMM)29 probabilistic modelling. DMM modeling was selected as the means for identifying community clusters due to the algorithm’s ability to generate mixture component vectors based on unique hyperparameters in a multinomial fashion. By design, this methodology intrinsically incorporates dynamic features with ranging sample sizes and species rareity when clustering communities of similar composition, therein making it an optimal tool for this meta-analysis.
Square root scaled GO term counts and taxonomic feature count matrices subjected to unsupervised community typing with DMM clustering (Suppl. Table 5) were subsequently compared by analysis of variance (ANOVA) with metadata categories case type and survival outcome. Statistically significant GO terms results derived from the MaAsLin2 analysis were thereafter ordered by parental lineage and visualized alongside consensus DMM clusters and metadata categories publication, case type, and survival outcome using the package pheatmap (v1.0.12)30.
Data and code availability
Overview of the processing workflow as well as all code used in the execution of the processing pipeline, analysis and visualization R scripts, and intermediate files have been made publicly available can be found online at the COV-IRT microbial GitHub repository (https://github.com/COV-IRT/microbial) and Open Science Framework (OSF) project (https://osf.io/7nrd3/) websites. Additional information about the commands and versions of the tools used to process raw reads and assign taxonomies and GO terms can be found online on the OSF project website (https://osf.io/7nrd3/).
Results
Comparison between subject categorical classes (i.e., uninfected controls, or patients with CAP or COVID-19 disease)
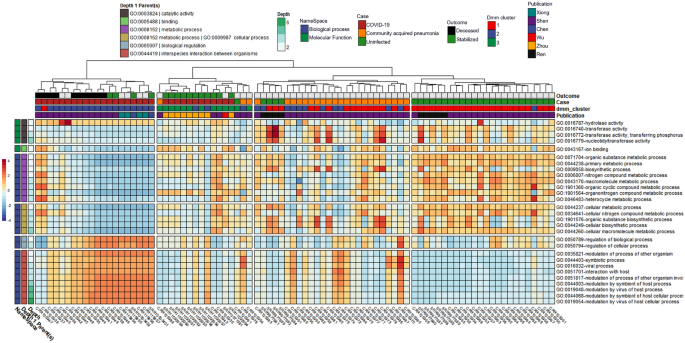
After controlling for random effects of publication and patient, results from the MaAsLin2 comparison across individual subjects were grouped by one of three categorical classes: (1) uninfected controls; (2) CAP patients; or (3) COVID-19 patients with moderate to severe disease, including death (Table 1). This revealed 20 out of 13,534 GO terms associated with COVID-19 when compared to the CAP cohort and 30 out of 13,534 GO terms associated with COVID-19 when compared to the uninfected cohort (Fig. 1, Tables 1 and 2). Significant GO terms were grouped under seven parental GO terms, including catalytic activity [GO:0003824], binding [GO.0005488], metabolic process [GO:0008152], cellular process [GO:0009987], biological regulation [GO:0065007], viral process [GO:0016032], and interspecies interaction between organisms [GO:0044419] (Fig. 1, Suppl. Table 3). Parental GO terms have smaller depth numbers (e.g., depth = 1) in the Gene Ontology hierarchy and represent higher-level features under molecular function [GO:0003674] or biological process [GO:0008150], whereas larger depth numbers represent nodes in the ontology tree that are lower and refer to more specific functions or processes.
Heatmap with notable microbially-derived gene ontology functional annotations associated with COVID-19 (n =32), as compared to community acquired pneumonia (n = 29) & uninfected (n=25) cohorts. Cells are colored via z-scale calculations of the total read counts for each GO term by sample. Rows are sorted by parental GO terms (depth = 1), and columns are clustered by Euclidean distance using ward D2 clustering. Comparisons were conducted using MaAsLin2, controlling for publication and patient ID with Benjamini Hochberg multiple test comparison (q < 0.05).
GO terms enriched in the COVID-19 cohort compared to the uninfected cohort included hydrolase activity [GO:0016787], as well as all significant GO terms with the parental terms of biological regulation [GO:0065007], viral process [GO:0016032], and interspecies interaction between organisms [GO:0044419]. Hydrolase activity [GO:0016787], nucleic acid metabolic process [GO:0090304], and many GO terms classified under interspecies interaction between organisms [GO:0044419] were also enriched in the COVID-19 cohort when compared to CAP. In contrast, GO terms enriched in the uninfected cohort compared to the COVID-19 cohort included all significant GO terms with the parental terms of cellular process [GO:0009987], metabolic process [GO:0008152], binding [GO.0005488], and terms classified under catalytic activity [GO:0003824] other than hydrolase activity [GO:0016787]. Results from the Dirichlet multinomial mixtures clustering analysis using all 13,534 gene ontology counts resulted in a best model fit using three distinct clusters that were significantly associated with each case cohort [p < 0.0001] (Fig. 1, Suppl. Table 5).
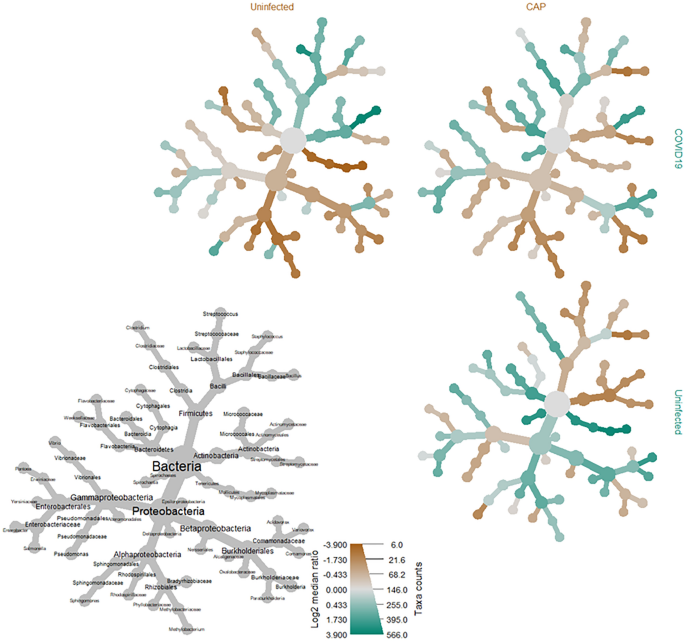
Taxonomic comparisons of the COVID-19 cohort to uninfected and CAP cohorts revealed 233 and 61 significantly differentiated species-level taxa with absolute values of log2 median ratios > 1.0 when comparing the COVID-19 cohort to uninfected and CAP cohorts, respectively (Suppl. Table 6). All significant taxa found in the CAP cohort were depleted compared to the COVID-19 cohort. Additionally, all significant taxa found in the CAP to COVID-19 comparison were also identified as significant in the uninfected to COVID-19 comparison (Suppl. Table 6). Of the taxa identified when comparing the uninfected cohort to the COVID-19 cohort, a total of 36 species were only marginally enriched (Suppl. Table 6).
Taxonomic comparisons resulted in a statistically significant difference amongst several microbial genera within the phylum of Proteobacteria, including those of the families Sphingomonadaceae (i.e., Sphingobium, Sphingopyxis, Sphingomonas) and Rhodobacteraceae (i.e., Paracoccus) when comparing the COVID-19 cohort to the uninfected (p < 0.001, q < 0.001) and CAP (p = 0.0067, q = 0.024) cohorts (Fig. 2, Table 4). There was a significant increase in several species belonging to the genus Sphingomonas among BALF specimens from COVID-19 patients when compared to both the uninfected (p < 0.0001, q < 0.001) and CAP cohorts (p < 0.005, q < 0.05) (Suppl. Table 6), with a more significant increase of Sphingomonas in COVID-19 patients when compared to the uninfected cohort than to the CAP cohort (Fig. 2). An analysis of the most common SeqScreen outputs taxonomically classified as Sphingomonas in BALF specimens among patients with COVID-19, irrespective of disease outcomes, included GO term assignments of hydrogen peroxide catabolic process [GO:0042744], response to oxidative stress [GO:0006979], catalase activity [GO:0004096], heme binding [GO:0020037], and metal ion binding [GO:0046872].
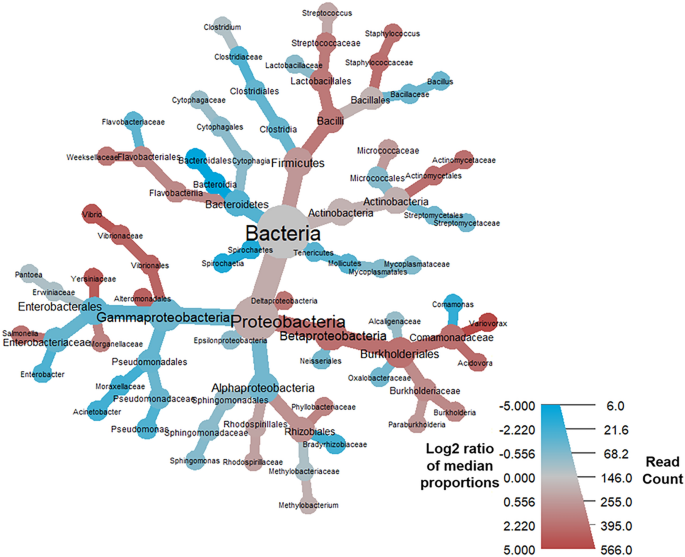
Heat tree matrix visualizing distinct COVID-19 vs. uninfected & community acquired viral pneumonia taxonomic profiles. This taxonomic heat tree data matrix visualization depicts the log2 median ratio differences across the three different cohorts. The tree depicted in grey in the lower left represents a taxonomic assignment key for all the other trees. Each of smaller trees represents a comparison between the different cohorts, as labelled in the columns and rows. The taxa colored brown are more abundant among the cohort labelled in the columns, whereas taxa colored green are more abundant in the cohort labelled in the rows. For example, there were significant increases (green) identified in log2 median ratio of several species belonging to the genus Sphingomonas when comparing the COVID-19 to the uninfected cohort (top left) and decreases (brown) when comparing the uninfected cohort to the community acquired pneumonia (CAP) cohort (bottom right).
There were no significant differences in alpha diversity when comparing case type (i.e., COVID-19, CAP, uninfected) (p-value = 0.051) or mortality (p-value = 0.8918) using the Shannon and inverse Simpson indices31,32. A full list of diversity metric indices is available in Supplementary Table 7. Beta diversity analyses did not reveal any statistically significant within group differences (F = 0.293, p = 0.747) by cohorts, which were determined by analysis of variance homogeneity of multivariate dispersions based on Euclidean distance. Further, no significant differences were observed in beta diversity amongst case type (F = 2.9257, p > 0.05) or mortality (F = 3.5978, p > 0.05), as determined by the permutation test for adonis using bray Curtis dissimilarity indices after stratifying by publication and patient.
Metatranscriptomic comparison of BALF specimens from COVID-19 subjects sub-categorized and stratified by disease survival or death
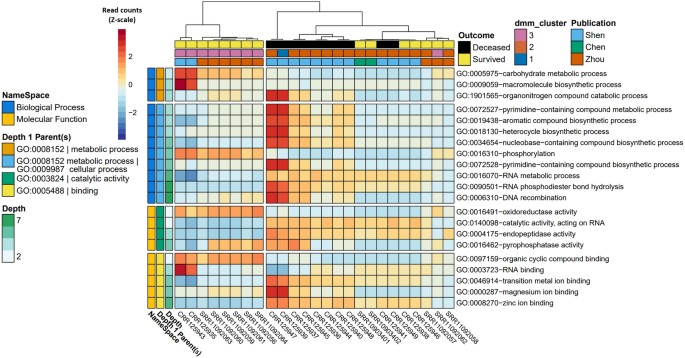
From subjects with known COVID-19 survival outcomes (i.e., of 32 samples, n = 10 deceased, and n = 15 survived), a stratified analysis amongst the categorical class of COVID-19 disease was performed via MaAsLin2. After controlling for random effects of patient, we observed 21 unique GO terms which were significantly increased in their association with death (n terms = 16, q-value < 0.05) or survival (n terms = 5, q-value < 0.05) from COVID-19 disease, with parental GO terms (depth = 1) of metabolic process [GO:0090304], binding [GO.0005488], and catalytic activity [GO:0003824] (Table 5, Fig. 3). GO terms with significant q-values (< 0.05) that were terminal in the observed GO term lineage (i.e., as specific as possible within the lineages of our result set), included nucleobase-containing compound biosynthetic process [GO:0034654], organonitrogen compound catabolic process [GO:1901565], pyrimidine-containing compound biosynthetic process [GO:0072528], and DNA recombination [GO:0006310] classified under the parental GO term of metabolic process [GO:0008152]; RNA binding [GO:0003723], magnesium ion binding [GO:0000287], and zinc ion binding [GO:0008270] classified under the parental GO term of binding [GO.0005488]; and oxidoreductase activity [GO:0016491] and endopeptidase activity [GO:0004175] classified under the parental GO term of catalytic activity [GO:0003824] (Suppl. Tables 4, 9–17).
Heatmap of significantly different gene ontology terms associated with COVID-19 mortality comparing deceased (n = 10) versus survived (n = 15). Cells are colored via z-scale calculations of the total read counts by sample (x axis) and by GO term (y axis). Rows are sorted by parental GO terms (depth = 1) and columns are clustered by Euclidean distance using ward D2 clustering. Comparisons were conducted using MaAsLin2, controlling for patient ID with Benjamini Hochberg multiple test comparison (q < 0.05).
Of the nine terminal GO terms that were significantly different in this analysis (q-value < 0.05), RNA binding [GO:0003723] and oxidoreductase activity [GO:0016491] were the most enriched in samples from individuals that survived COVID-19 (Suppl. Table 4). An analysis of the proteins underlying the SeqScreen GO term assignments showed that RNA binding [GO:0003723] was driven by an enrichment of 30S and 50S ribosomal proteins from the Gram-positive cocci belonging to the genera Streptococcus, Granulicatella, Enterococcus, and Lactococcus, all of which were particularly prevalent in the “nCov7” survived COVID-19 patient from the Shen et al. study (Suppl. Table 8). The enrichment of the oxidoreductase activity [GO:0016491] term among survived COVID-19 patients was driven by many different samples and a variety of bacteria, including those from Gram-positive bacteria belonging to the genera Enterococcus, Streptococcus, Streptomyces, Pediococcus, Lactococcus, and Granulicatella. Examples of underlying reference proteins to which reads mapped resulting in our observed oxidoreductase activity [GO:0016491] term included quinone oxidoreductase, pyruvate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, and glyceraldehyde-3-phosphate dehydrogenase (Suppl. Table 14). Among the deceased COVID-19 patients, the terminal GO terms of endopeptidase activity [GO:0004175], zinc ion binding [GO:0008270], and nucleobase-containing compound biosynthetic process [GO:0034654] were being driven by an enrichment of SARS-CoV-2 proteins (Suppl. Tables 10, 12, 14). Mixed among proteins from other organisms, an enrichment of Variovorax proteins tagged with the terminal GO terms of pyrimidine-containing compound biosynthetic process [GO:0072528] (e.g., CTP synthase, putative sulfonate/nitrate transport system substrate-binding protein), organonitrogen compound catabolic process [GO:1901565] (e.g., histidine ammonia-lyase, aspartate/glutamate leucyltransferase), magnesium ion binding [GO:0000287] (e.g., proteins involved in the histidine biosynthesis pathway, such as phosphoribosyl-AMP cyclohydrolase), and DNA recombination [GO:0006310] (e.g., inclusive of possible Variovorax phage proteins—integrase family protein, putative transposase IS4 family, phage integrase family protein) appeared in the COVID-19 deceased patients. This enrichment of Variovorax proteins among samples from individuals who died of COVID-19 was consistent with the results from the taxonomic comparison analysis. Compared to the survived group, the taxonomic comparisons in the deceased group revealed a statistically significant (p < 0.0001, q < 0.001) increase of the family Comamonadaceae, belonging to the genus Variovorax, and decreases in the family Bacteriodales (Fig. 4, Table 6).
Heat tree demonstrating the BALF metatransciptome profiles associated with COVID-19 mortality. Taxa colored in red were more prevalent amongst COVID-19 patients who died, and nodes in blue represent taxa that were more prevalent amongst patients who survived COVID-19. Notable increases were observed in the log2 median ratios in the family Comamonadaceae, genus Variovorax, and significant decreases in the log2 median ratios of order Bacteroidia and class Bacteroidales.
Discussion
We observed significantly unique discriminant taxonomic and functional features in bronchoalveolar lavage fluid (BALF) metatranscriptomes in association with COVID-19 disease and its mortality. Of note, due to limitations in the depth of clinical metadata by subject, we could not distinguish between COVID-19 pathophysiology or associated medical comorbidities, treatments, nor interventions. However, because of the time interval in which COVID-19 patient specimens were recruited to their respective index studies at the beginning of the outbreak in Wuhan, China (i.e., 2019 and early 2020), COVID-19-specific interventions and treatments had yet to be introduced and thus comparisons between CAP and COVID-19 subject specimens would be less likely to be related to disease-focused therapy.
Results driven by coronavirus protein functions
At the time of this study, the standard Kraken2 taxonomic database included the SARS-CoV-2 reference genome, but the SARS-CoV-2 proteins were not yet added to the SeqScreen database that was used for the functional analysis. This functional analysis demonstrated how GO terms and their corresponding proteins can be used to characterize an emerging pathogen (i.e., a pathogen that is not present in the reference database), as well as significant host microbiome functional shifts. SARS-CoV-2 reads were successfully detected in the taxonomic analysis of COVID-19 BALF samples, and GO terms associated with coronavirus proteins were found to be significantly different in the functional analysis. A number of coronavirus proteins were driving the significant associations of GO terms between COVID-19 and uninfected samples, including modulation by symbiont of host cellular process [GO:0044068], modulation by virus of host cellular process [GO:0019054], modulation by virus of host process [GO:0019048], modulation of process of other organism involved in symbiotic interaction [GO:0051817], modulation by symbiont of host process [GO:0044003], interaction with host [GO:0016032], viral process [GO:0051701], interspecies interaction between organisms [GO:0044419], modulation by symbiont of host cellular process [GO:0044068], and modulation by virus of host cellular process [GO:0019054] (Suppl Table 3). Coronavirus proteins were also driving notable GO term associations in COVID-19 deceased vs. survived, including transition metal ion binding [GO:0046914], zinc ion binding [GO:0008270], organic cyclic compound binding [GO:0097159], endopeptidase activity [GO:0004175], and nucleobase containing compound biosynthetic process [GO:0034654]. While samples from both COVID-19 deceased and survived individuals contained taxonomically and functionally classified coronavirus reads, the significant terminal GO terms of endopeptidase activity [GO:0004175], zinc ion binding [GO:0008270], and nucleobase-containing compound biosynthetic process [GO:0034654] were positively correlated with COVID-19 deceased patients. This was likely due to multiple highly expressed coronavirus proteins being tagged with these GO terms (e.g., replicase polyprotein 1ab, 2'-O-methyltransferase), and a higher SARS-CoV-2 viral load and mRNA expression being present in patients who died of COVID-19 disease.
Significant taxonomic differences observed in the microbial communities
Distinct taxonomic features of BALF specimens from the COVID-19 vs. uninfected analysis included an increase in the genus Sphingomonas, belonging to the Sphingomonadacae family, among COVID-19 patients. Notable taxonomic features among COVID-19 patients with mortal disease included increases in log2 median ratios of the genus Variovorax, belonging to the Comamonadaceae family, and decreases in the class Bacteroidia, belonging to the order Bacteroidales. These findings support previous reports regarding an association with Gram-negative Sphingomonas33,34,35,36, which is a common opportunistic pathogen found in nosocomial infections. A previous 16S rRNA profiling study by Gaibani et al. found that the BALF of critically ill COVID-19 patients had lower amounts of commensal bacterial species and an enrichment of opportunistic Gram-negative pathogens, which was often associated with multidrug resistance40. Among the COVID-19 cohort, one of the most highly expressed Sphingomonas genes was catalase [UniProt ID = J8VPL9]. This Sphingomonas catalase protein is assigned GO terms including hydrogen peroxide catabolic process [GO:0042744], response to oxidative stress [GO:0006979], catalase activity [GO:0004096], heme binding [GO:0020037], and metal ion binding [GO:0046872], and it is responsible for decomposing hydrogen peroxide into water and oxygen. This serves to protect cells from the toxic effects of hydrogen peroxide, which may suggest that Sphingomonas spp. respond to COVID-19 conditions in the patient by expressing genes that help them to survive well in environments undergoing great amounts of oxidative stress.
Our findings additionally support a previous report regarding an increase in the abundance of Variovorax in COVID-19 patient BALF tissue37. Variovorax spp. Have also previously been reported in the microbiota of patients with lung cancer38 and were shown to be a key driver of clustering amongst patients challenged with H1N1 influenza infections39. The most abundantly expressed Variovorax proteins in the COVID-19 cohort included those involved in cell wall organization and the plasma membrane (e.g., binding-protein-dependent transport systems inner membrane component [UniProt ID = E6VB76], endolytic peptidoglycan transglycosylase RlpA [UniProt ID = T1XG48]), oxidoreductase activity (e.g., methylenetetrahydrofolate reductase [UniProt IDs = J2L4W7, T1XH55], taurine dioxygenase [UniProt ID = T1XBI4], NADH-quinone oxidoreductase subunit H [UniProt ID = E6V509]), hydrolase activity (e.g., N-acyl-d-aspartate/d-glutamate deacylase [UniProt ID = J2T0U3], cytokinin riboside 5'-monophosphate phosphoribohydrolase [UniProt IDs = E6V0P4, J3CLH3]), and ATP-binding transport (e.g., ABC transporter related protein [UniProt ID = E6UUY9], extracellular solute-binding protein family 5 [UniProt ID = E6V3F7]).
These findings of this study are consistent with a prior 16S rRNA profiling study by Bassis et al., where BALF from healthy subjects was found to contain bacteria from the genera Prevotella (class Bacteroidia), Veillonella, and Streptococcus41. In addition to the significance of the RNA binding GO term [GO:0003723] being driven by an enrichment of 30S and 50S ribosomal proteins from Gram-positive cocci like Streptococcus in the survived COVID-19 cohort (Suppl. Table 8), the endopeptidase activity GO term [GO:0004175] was connected to membrane organization proteins for Gram-positive bacteria (e.g., Gram-positive signal peptide protein, YSIRK family), which were more prevalent in the COVID-19 survived cohort. This study also found the class Bacteroidia to be increased in the survived COVID-19 cohort.
Enrichment of the histidine biosynthesis pathway
The genes underlying the significant GO term magnesium ion binding [GO:0000287] revealed an enrichment of transcripts involved in microbial biosynthesis pathways in the COVID-19 deceased cohort (e.g., Phosphoribosyl-AMP cyclohydrolase). Prior experiments have found that histidine biosynthesis is critical for the pathogen Klebsiella pneumoniae to grow in immunosuppressed lungs42, and histidine serves as a crucial nitrogen source for infections by the nosocomial pathogen Acinetobacter baumannii43. For these reasons, it was proposed that the histidine biosynthesis pathway could be a promising drug target to combat opportunistic bacterial infections42,43. In this study, the enrichment of gene transcripts within the histidine biosynthesis pathway among the COVID-19 deceased cohort suggests that histidine could be an important contributor to the survival and pathogenicity of opportunistic bacteria in the BALF of COVID-19 patients.
Evidence of stress responses in bacterial pathogens
Several of the enriched gene transcripts identified in this study were involved in different bacterial stress response pathways. The zinc ion binding GO term [GO:0008270] was enriched in SARS-CoV-2 proteins, but it was also connected to an increased expression of genes involved in the formaldehyde bacterial stress response in COVID-19 deceased individuals (e.g., S-(hydroxymethyl)glutathione dehydrogenase, Glutathione-independent formaldehyde dehydrogenase). Formaldehyde is highly toxic to microbes, and this study showed evidence of genes within the most widespread pathway for formaldehyde detoxification44 (where thiol in tripeptide glutathione serves as the initial formaldehyde acceptor) to be enriched in the COVID-19 deceased cohort. Also enriched in the COVID-19 deceased cohort were genes labeled with the DNA recombination GO term [GO:0006310] and involved in phage activity (e.g., Variovorax phage proteins—integrase family protein, putative transposase IS4 family, phage integrase family protein). Prophage activities have been previously shown to contribute to the survival and pathogenicity of bacteria and may be activated in response to stress45,46,47,48. The enrichment of the oxidoreductase activity GO term [GO:0016491] among the COVID-19 survived cohort included underlying genes such as quinone oxidoreductase, pyruvate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, and glyceraldehyde-3-phosphate dehydrogenase. Lung disease may become more severe in COVID-19 with increased oxidative stress, and it is possible that bacterial response in the COVID-19 survived cohort helped to reduce the oxidative stress49,50,51.
Conclusions
COVID-19 disease has demonstrated a wide range of clinical severity outcomes, but the factors that correlate with disease severity are not fully understood. Here we identified significant taxonomic and functional differences in BALF metatranscriptomes associated with COVID-19 disease and death. More significant differences were observed between the COVID-19 disease and uninfected cohorts than the COVID-19 disease and CAP cohorts, suggesting correlations specific to SARS-CoV-2 infection. Significant differences were also found associated with COVID-19 mortality. Discriminant taxonomic differences associated with COVID-19 disease and mortality included the following: the genus Sphingomonas significantly increased with COVID-19 disease compared to the uninfected cohort and to a lesser extent with COVID-19 disease compared to the CAP cohort, the genus Variovorax significantly increased with COVID-19 mortality, and in the class Bacteroidia significantly decreased with COVID-19 mortality. Compared to the patients who were reported to have survived COVID-19 disease, the metatranscriptome data from COVID-19 deceased individuals showed a significant increase in specific GO terms assigned to SARS-CoV-2 proteins, which was likely because of their higher SARS-CoV-2 viral load. Additionally, COVID-19 deceased individuals showed more transcripts from genes involved in the histidine biosynthesis pathway and demonstrated evidence of active bacterial stress response pathways. By the nature of this analysis, this work does not address causality or directionality. However, this work does identify a relationship between the human microbiome and COVID-19 morbidity and mortality, and the specific functions and taxa identified warrant further investigation. Although this experiment was focused on the impact of COVID-19 disease on the BALF microbiome, none of the methods employed in this study were specific to COVID-19. We hope that the methods implemented here will be useful to the research community for other microbiome and pathogen-related association experiments, particularly as more metatranscriptome sequences and pathogenesis gene ontologies are created in the future52,53.
Data availability
The original sequence datasets used in this study were previously published and are publicly available in the locations described in Suppl. Tables 1 and 2. An overview of the data processing workflow, all code used in the execution of the processing pipeline, analysis and visualization R scripts, and intermediate files have been made publicly available can be found online at the COV-IRT microbial GitHub repository (https://github.com/COV-IRT/microbial) and Open Science Framework (OSF) project (https://osf.io/7nrd3/; doi: 10.17605/OSF.IO/7NRD3) websites. The OSF wiki (https://osf.io/7nrd3/wiki/home/) describes specific software tools and commands that were used to generate the results. The OSF project includes the following high-level project components relevant to this manuscript: Microbial_Pre-Processing (i.e., outputs from quality trimming and filtering of raw sequence data), Metatranscriptome_Kraken2 (i.e., Kraken2 taxonomic classification outputs), Metatranscriptome_SeqScreen (i.e., SeqScreen final reports), Metatranscriptome_GO_Terms and Metatranscriptome_GO_Term_Summaries (i.e., summaries of SeqScreen-assigned GO terms), and Metatranscriptome_UniProt_ID_Counts (i.e., summaries of SeqScreen-assigned UniProt IDs). All methods were carried out in accordance with relevant guidelines and regulations. Suppl. Table 18 provides legends for all supplementary tables.
References
Merenstein, C. et al. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. MBio 12(4), e0177721 (2021).
Shen, Z. et al. Genomic diversity of severe acute respiratory syndrome—Coronavirus 2 in patients with coronavirus disease 2019. Clin. Infect. Dis. 71(15), 713–720 (2020).
Haiminen, N., Utro, F., Seabolt, E. & Parida, L. Functional profiling of COVID-19 respiratory tract microbiomes. Sci. Rep. 11(1), 6433 (2021).
Yang, H., Zhilong, J., Jinlong, S., Weidong, W. & Kunlun, H. The active lung microbiota landscape of COVID-19 patients through the metatranscriptome data analysis. BioImpacts (BI) 12(2), 139–146 (2021).
Chen, L. et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 9, 313–319 (2020).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Xiong, Y. et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 9, 761–770 (2020).
Michalovich, D. et al. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat. Commun. 10, 5711 (2019).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181(5), 1036-1045.e9 (2020).
Daamen, A. R. et al. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 11(1), 7052 (2021).
Abouelkhair, M. A. Non-SARS-CoV-2 genome sequences identified in clinical samples from COVID-19 infected patients: Evidence for co-infections. PeerJ 8, e10246 (2020).
Khan, A. A. & Khan, Z. COVID-2019-associated overexpressed Prevotella proteins mediated host-pathogen interactions and their role in coronavirus outbreak. Bioinformatics 36(13), 4065–4069 (2020).
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Huang, W. et al. Optimizing a metatranscriptomic next-generation sequencing protocol for bronchoalveolar lavage diagnostics. J. Mol. Diagn. 21, 251–261 (2019).
Ren, L. et al. Transcriptionally active lung microbiome and its association with bacterial biomass and host inflammatory status. mSystems. 30, 199 (2018).
Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 2114–2120 (2014).
Magoč, T. & Salzberg, S. L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 1–13 (2019).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Balaji, A. et al. Accurate and sensitive functional screening of pathogenic sequences via ensemble learning. Genome Biol. 23(1), 133 (2022).
Lee, M. bit: A multipurpose collection of bioinformatics tools. F1000Research 11, 122. https://doi.org/10.12688/f1000research.79530.1 (2022).
McMurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
R Core Team. R: A Language and Environment for Statistical Computing. https://www.R-project.org/ (R Foundation for Statistical Computing, 2021).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 6, 1–14 (2018).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17(11), e1009442 (2021).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological) 57(1), 289–300 (1995).
Foster, Z. S. L., Sharpton, T. J. & Grünwald, N. J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PloS Comput. Biol. 13, e1005404 (2017).
Holmes, I., Harris, K. & Quince, C. Dirichlet multinomial mixtures: Generative models for microbial metagenomics. PLoS ONE 7, e30126 (2012).
Kolde, R. pheatmap: Pretty Heatmaps. https://cran.r-project.org/web/packages/pheatmap/ (2018).
Shannon, C. E. A mathematical theory of communication. Bell Syst. Tech. J. 27(3), 379–423 (1948).
Simpson, E. H. Measurement of diversity. Nature 163(4148), 688 (1949).
Sirivongrangson, P. et al. Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med. Exp. 8, 72 (2020).
Chen, S. et al. Clinical and etiological analysis of co-infections and secondary infections in COVID-19 patients: An observational study. Clin. Respir. J. 15, 815–825 (2021).
Ryan, M. P. & Adley, C. C. Sphingomonas paucimobilis: A persistent Gram-negative nosocomial infectious organism. J. Hosp. Infect. 75, 153–157 (2010).
Hsueh, P. R. et al. Nosocomial infections caused by Sphingomonas paucimobilis: Clinical features and microbiological characteristics. Clin. Infect. Dis. 26, 676–681 (1998).
Han, Y., Jia, Z., Shi, J., Wang, W. & He, K. The active lung microbiota landscape of COVID-19 patients through the metatranscriptome data analysis. BioImpacts (BI). 12(2), 139–146 (2021).
Rose, U. D. et al. Role of the microbiota in primary lung cancer initiation and progression. J. Immunol. 202, 1901–1901 (2019).
Chaban, B. et al. Characterization of the upper respiratory tract microbiomes of patients with pandemic H1N1 influenza. PLoS ONE 2013, 8 (2013).
Gaibani, P. et al. The lower respiratory tract microbiome of critically ill patients with COVID-19. Sci. Rep. 11, 10103 (2021).
Bassis, C. M. et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6(2), e00037 (2015).
Silver, R. J. et al. Amino acid biosynthetic pathways are required for Klebsiella pneumoniae growth in immunocompromised lungs and are druggable targets during infection. Antimicrob. Agents Chemother. 63(8), e02674-e2718 (2019).
Lonergan, Z. R., Palmer, L. D. & Skaar, E. P. Histidine utilization is a critical determinant of Acinetobacter pathogenesis. Infect. Immun. 88(7), e00118-20 (2020).
Chen, N. H., Djoko, K. Y., Veyrier, F. J. & McEwan, A. G. Formaldehyde stress responses in bacterial pathogens. Front. Microbiol. 7, 257 (2016).
Matos, R. C. et al. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PloS Genet. 9(6), e1003539 (2013).
Wagner, P. L. & Waldor, M. K. Bacteriophage control of bacterial virulence. Infect. Immun. 70(8), 3985–3993 (2002).
Wang, X. et al. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1, 147 (2010).
Carey, J. N. et al. Phage integration alters the respiratory strategy of its host. Elife 8, e49081 (2019).
Derouiche, S. Oxidative stress associated with SARS-Cov-2 (COVID-19) increases the severity of the lung disease—A systematic review. Infect. Dis. Epidemiol. 6, 121 (2020).
Wieczfinska, J., Kleniewska, P. & Pawliczak, R. Oxidative stress-related mechanisms in SARS-CoV-2 infections. Oxid. Med. Cell. Longev. https://doi.org/10.1155/2022/5589089 (2022).
Seixas, A. F. et al. Bacterial response to oxidative stress and RNA oxidation. Front. Genet. 12, 821535 (2022).
Godbold, G. D., Kappell, A. D., LeSassier, D. S., Treangen, T. J. & Ternus, K. L. Categorizing sequences of concern by function to better assess mechanisms of microbial pathogenesis. Infect. Immun. 90(5), 33421 (2022).
Pathogenesis Gene Ontology (PathGO). GitHub. https://github.com/jhuapl-bio/pathogenesis-gene-ontology (2022).
Acknowledgements
We would like to thank the COVID-19 International Research Team (COV-IRT) microbial subgroup team members and give special acknowledgment to John Fonner and the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported. We would also like to acknowledge the thoughtful review and suggestions provided by Dr. Enrico R. Barrozo, which led to improvements in our manuscript.
Funding
Dr. Michael Jochum was supported by Grant Number T32 HD098069 from NIH NICHD. Dr. Treangen was supported in part by the National Institute of Allergy and Infectious Diseases (Grant# P01-AI152999). The research performed by Drs. Ternus and Treangen for this study was partially funded by the Fun GCAT program from the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via the Army Research Office (ARO) under Federal Award No. W911NF-17-2-0089. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the ODNI, IARPA, ARO, or the US Government.
Author information
Authors and Affiliations
Contributions
M.J. statistically analyzed the taxonomic and functional results, created the figures, and wrote the manuscript. M.D.L. developed code for sequence data analysis and provided reviews and guidance on how to analyze the data and generate figures. K.C. led the effort to run SeqScreen on computational resources provided at the Texas Advanced Computing Center. V.Z. reviewed data analysis results, performed background literature reviews, and contributed to writing the manuscript. E.V. assisted in analyzing the functional results and reviewing results in the context of prior studies. T.T. provided guidance in experimental design and computational analyses, as well as contributing to researching relevant prior publications. K.A. provided clinical expertise and reviewed the experimental design, data analysis results, and conclusions for clinical accuracy and relevance. K.L.T. oversaw and contributed to the experimental design, data analysis, and writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jochum, M., Lee, M.D., Curry, K. et al. Analysis of bronchoalveolar lavage fluid metatranscriptomes among patients with COVID-19 disease. Sci Rep 12, 21125 (2022). https://doi.org/10.1038/s41598-022-25463-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25463-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.