Abstract
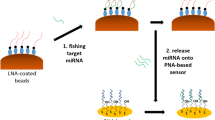
Abnormal expression levels of miRNA are associated with various tumor diseases, for example, glioma tumors are characterized by the up-regulation of miRNA-182. Surface plasmon resonance (SPR) assay for miRNA-182 from glioma patients was performed via DNA walking amplification strategy. The duplex between aminated swing arm DNA (swDNA) and block DNA (blDNA), and aminated track DNA (trDNA) with a biotin tag were tethered on the poly(ethylene glycol) (PEG)-modified chips. Upon formation of miRNA/blDNA duplex, the SPR signal decreased with the walking process of swDNA, as the biotinylated fragment of trDNA (biotin-TTGGAGT) was detached from the sensor surface caused by the nicking endonuclease Nb.BbvCI. Such a repeated hybridization and cleavage cycle occurred continuously and the detachment of more biotinylated fragments of trDNA from the chips led to the attachment of fewer streptavidin (SA) molecules and then smaller SPR signals. MiRNA-182 with concentrations ranging from 5.0 fM to 1.0 pM could be readily determined and a detection limit of 0.62 fM was achieved. The proposed method was highly selective and possessed remarkable capability for evaluating the expression levels of miRNA-182 in serum samples from healthy donors and glioma patients. The sensing protocol holds great promise for early diagnosis of cancer patients.
Similar content being viewed by others
Introduction
As a kind of non-coding and single-stranded RNAs with 18–25 nucleotides, microRNAs (miRNAs) participated in the regulation of gene expression in a large number of biological processes. Abnormal miRNA expression was closely related to cancers1,2,3, cardiovascular diseases4, and neurological diseases5. However, the detection of miRNAs was challenged by low expression level, short sequence length, ease of degradation, and high sequence similarity.
Traditional methods for miRNA assay included but not limited to microarray6, northern blotting7, and quantitative real-time polymerase chain reaction (qRT-PCR)8. However, these methods either suffered from low sensitivity, or possessed the drawbacks, such as time consumption, and operation complexity. To overcome these problems, the strategies for miRNA assay based on surface plasmon resonance (SPR)9,10,11,12,13,14,15,16,17,18,19,20,21,22,23, electrochemistry24,25,26,27, fluorescence28,29,30, electrochemical luminescence31,32,33, and colorimetry34,35,36 have emerged. As an optical technique, SPR is capable of measuring the changes in refractive index or thickness occurring at the surface of a metal film (typically gold)37, being advantageous over other techniques due to its label-free, real-time, and highly sensitive properties37. Typically, the SPR signal can be amplified through recognition by antibodies9 and nanoparticles10,12,13, or biotin-streptavidin complexation10,14, catalytic hairpin assembly15,16,17, and enzymatic reaction18. Metal nanoparticles have been widely applied for SPR detection of miRNA. For example, based on DNA amplification and adsorption of silver nanoparticles (AgNPs), enzyme-free SPR assay of miRNA with a detection limit of 0.6 fM has been carried out11. In situ generated AgNPs via hybridization chain reaction were involved for amplified SPR detection of let-7a miRNA12. Wang et al. have proposed a SPR platform for miRNA-21 assay by coupling gold nanoparticles (AuNPs) with super sandwich assembly19. Furthermore, the same group constructed graphene oxide-AuNPs hybrids for SPR detection of miRNA-141 extracted from human cancer cell lines20. Although the metal nanoparticles with high molecular weight and the electronic coupling between metal nanoparticles and the Au film could effectively enhance the SPR signals38,39, the nonspecific adsorption of metal nanoparticles limited their further applications in SPR. SA is widely used to amplify the SPR signals due to its large molecular weight (56 kDa) and strong binding with biotin (KD = 10–15 M−1) 40.
DNA nanotechnology-based systems, such as HCR41, rolling circle amplification (RCA)42, and DNA walking systems43 have emerged for biosensing due to easy fabrication, low cost, and good stability44. The DNA walking system consisting of a swing arm, a track, and a trigger has attracted much attention45. Typically, the target triggered the DNA walking mechanism and the swing arm moved automatically along the track, leading to cascade signal amplification. Toehold-mediated strand displacement reaction (TSDR) served as a promising alternative for detection of single-nucleotide polymorphisms due to its high selectivity46,47. To perform highly sensitive and specific SPR assay, combining TSDR, DNA walking process and enzymatic cleavage reaction might be a better choice.
Herein, sensitive and selective detection of miRNA was achieved using SPR. In the presence of miRNA-182, the SPR signal decreased with the walking process, which was caused by the detachment of the biotinylated fragments of trDNA (biotin-TTGGAGT) from the sensor surface with the help of Nb.BbvCI. The proposed method holds great promise for early diagnosis of miRNA-related major diseases.
Experimental
Materials
Triethylene glycol mono-11-mercaptoundecyl ether (HSC11PEG3-OH), ethanolamine hydrochloride (EA), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS) and streptavidin (SA) were purchased from Sigma-Aldrich (St. Louis, MO). Hexaethylene glycol mono-11-mercaptoundecyl acid (HSC11PEG6-COOH) was acquired from Dojindo Laboratories (Kumamoto, Japan). Anhydrous ethanol, NaCl, MgCl2, K2HPO4, KH2PO4, agarose, DNA marker, 6× loading buffer, and diethylpyrocarbonate (DEPC)-treated deionized water were obtained from Sangon Biotech Co., Ltd. (Shanghai, China). The nicking endonuclease Nb.BbvCI was obtained from New England Biolabs (Ipswich, MA). High performance liquid chromatography (HPLC)-purified miRNAs and DNAs were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) and their sequences were listed in Table 1 (mismatched bases relative to that of miRNA-182 were underlined). Other reagents were of analytical purity and used as received. All solutions were prepared with deionized water treated with a water purification system (Simplicity185, Millipore Corp., Billerica, MA).
Procedures
Solution preparation
HSC11PEG6-COOH and HSC11PEG3-OH were dissolved in anhydrous ethanol. The mixed solution of 0.4 M EDC and 0.1 M NHS in deionized water was used to activate the carboxylic acid groups on PEG chips. The swDNA/blDNA duplex was formed by mixing swDNA and blDNA in phosphate-buffered saline (PBS, pH 7.4, 10 mM phosphate/10 mM NaCl) at a molar ratio of 1:2, heating to 95 °C for 5 min and then cooling down to room temperature. To maintain an RNase-free environment, the miRNAs were dissolved in PBS containing 5 mM MgCl2 that was prepared with DEPC-treated deionized water. The nicking endonuclease Nb.BbvCI was diluted with 1× CutSmart buffer (pH 7.9) containing 50 mM potassium acetate, 20 mM tris–acetate, 10 mM magnesium acetate and 100 μg/mL bovine serum albumin. SA was diluted to 50 nM with PBS.
Chip modification and SPR detection
The PEG-covered chips were formed by immersing the cleaned Au films in a mixed solution of HSC11PEG6-COOH (0.2 mM) and HSC11PEG3-OH (1.8 mM) in the dark for 48 h. Via activation of the carboxylic acid groups at the PEG chain end with EDC/NHS for 30 min at room temperature, the aminated swDNA/blDNA duplex and aminated trDNA with a molar ratio of 1:100 were anchored on the PEG chips through amide bond formation. After washing thoroughly with PBS and deionized water, the sensor chips were soaked in 0.1 M EA for 10 min to block the unreacted sites. Serum samples or miRNA with various concentrations were cast onto the chips for 3 h at 37 °C. After completion of the cleavage reaction by 50 U/mL of Nb.BbvCI for 2 h at 37 °C, the sensor chips were mounted onto the BI-4000 SPR instrument (Biosensing Instrument Inc., Tempe, AZ). Finally, 50 nM SA solution was preloaded into a 200 μL-sample loop and delivered to the sensor chips by the carrier solution at a flow rate of 20 μL/min. The degassed PBS that was treated by vacuum pumping for 30 min served as the carrier solution.
Agarose gel electrophoresis
Agarose gels (4%) were prepared as reported48. Briefly, 2.0 g agarose was dissolved in 50 mL 1× Tris‐acetate‐EDTA (TAE) buffer with the aid of microwave oven. Afterwards, 2.5 μL nucleic acid stain was evenly mixed with the slightly cooled agarose solution, followed by pouring into the gel mold. The analytes were loaded onto the gels in 6× DNA loading buffer. Gel electrophoresis was run in 1× TAE buffer at 120 V for 1 h on a Mini-Sub Cell GT electrophoresis system (Bio-Rad Laboratories, Hercules, CA), and imaged on a GelView 1500Plus intelligent gel imaging system (Guangzhou, China).
Results and discussion
The principle for SPR assay of miRNA based on DNA walking process and enzymatic cleavage reaction was depicted in Fig. 1. The aminated swDNA/blDNA duplex and aminated trDNA with a biotin tag were tethered on the PEG-modified chips through EDC/NHS coupling chemistry. The blDNA contained an exposed 9-nt toehold domain (orange color), which was complementary to the sequence of target miRNA. In the absence of miRNA-182, the biotin tag on the surface-confined trDNA remained intact and could be recognized by SA (56 KDa)40. Consequently, the large mass of SA amplified the SPR signals (a). However, the existence of miRNA triggered the TSDR, leading to the release of swDNA by forming blDNA/miRNA duplex. The released swDNA was then hybridized with the surface-confined trDNA and the biotinylated fragment on trDNA (biotin-TTGGAGT) could be cleaved by nicking endonuclease Nb.BbvCI with high sequence specificity (3′-GGAGT↓CG-5′)49,50,51. The released swDNA then autonomously walked on the chip surface, and with the help of Nb.BbvCI, more biotinylated fragments of trDNA were detached from the chips. As a result, the attachment of SA was remarkably hindered, leading to smaller SPR signals (b).
The occurrence of TSDR was confirmed by gel electrophoresis (Fig. 2A). The single band with different migration rates in lane 1, lane 2 and lane 4 corresponded to blDNA, swDNA and miRNA. After hybridization with blDNA, a new band with slower migration appeared, indicating the swDNA/blDNA duplex formation (top band in lane 3). The existence of miRNA triggered the walking mechanism, and the formation of blDNA/miRNA duplex was evidenced by a new band in lane 5 (surrounded by green circle). In the same time, the color of the band corresponding to swDNA deepened (middle band in lane 5), which demonstrates that the swDNA was successfully released. Gel electrophoresis assay of the DNA chains after enzymatic cleavage reaction was performed (Fig. S1). The single bands in lane 1 and lane 2 corresponded to trDNA and swDNA, respectively. Via hybridization between swDNA and trDNA, the bright band in lane 3 indicates the formation of the swDNA/trDNA duplex (lane 3). After the enzymatic cleavage reaction, the trDNA was broken into two fragments of TTTTTTTTTTGC (blue square, lane 4) and TGAGGTT (too small to be seen).
(A) Gel electrophoresis characterization of the TSDR. Lane 1: 2 μM blDNA; Lane 2: 1 μM swDNA; Lane 3: 1 μM swDNA/2 μM blDNA duplex; Lane 4: 2 μM miRNA-182; Lane 5: 1 μM swDNA/2 μM blDNA duplex + 2 μM miRNA-182. (B) SPR sensorgrams upon injection of 50 nM SA onto the Nb.BbvCI-treated fluidic channels that were covered with (a) trDNA + swDNA/blDNA duplex; (b) trDNA + swDNA/blDNA + miRNA-182; (c) trDNA + swDNA; (d) non-biotinylated trDNA + swDNA/blDNA duplex + miRNA-182.
Via combining DNA walking process and enzymatic cleavage reaction, SPR assay of miRNA was conducted (Fig. 2B). In the absence of miRNA, the protection of swDNA by blDNA hindered the occurrence of TSDR, and the recognition of the biotinylated trDNA by SA produced a large SPR signal of 104 mDeg (curve a, Fig. 2B). However, the existence of miRNA causes miRNA/blDNA duplex formation and the subsequent release of swDNA, triggered the walking mechanism. The released swDNA then hybridized with the surface-confined trDNA and the biotinylated fragment of trDNA could be cleaved by Nb.BbvCI. With the release of more swDNA, large numbers of trDNAs were enzymatically cleaved. As a result, the incorporation of SA led to an SPR signal of 41 mDeg (curve b, Fig. 2B). Note that the aminated swDNA/blDNA duplex and aminated trDNA were simultaneously anchored on the PEG-modified chips, and the molar ratio between swDNA/blDNA duplex and trDNA was optimized. The swDNA/blDNA duplex was first formed through hybridization of 25 nM swDNA with 50 nM blDNA. The resultant duplex was mixed with trDNA with different molar ratios, and then immobilized on the PEG chips. The SPR signals upon injection of 50 nM SA increased with the molar ratio (swDNA:trDNA) in the range of 1:20, 1:40, 1:80, 1:100 and 1:120, and began to level off beyond 1:100 (Fig. S2A). Furthermore, the extent of cleavage reached the maximum at 2 h (Fig. S2B). Thus, swDNA (25 nM)/blDNA (50 nM) duplex and 2.5 μM trDNA were tethered on the PEG chips, and the optimal enzymatic cleavage time was maintained at 2 h. The relatively large SPR signal of 41 mDeg caused by incomplete cleavage reaction (curve b) might be ascribed to the limited walking area of the released swDNAs on the two-dimensional sensor surface, as evidenced by the similar SPR signal in the case of unprotected swDNA and trDNA (curve c, Fig. 2B). When the biotinylated trDNAs in curve b were replaced by the non-biotinylated ones, almost no SPR signal was obtained upon injection of 50 nM SA (curve d, Fig. 2B), indicating that the nonspecific adsorption of SA on the chip surface was negligible.
The selectivity of the method for miRNA-182 assay was assessed by examining several miRNA-182 family members (miRNA-96, miRNA-182-3p)52,53 and other miRNA sequence (miRNA-155) (Fig. 3). The signal difference in the vertical axis in Fig. 3 was defined by subtracting the SPR signals in the case of different miRNAs from the blank (curve a in Fig. 2B). MiRNA-96, miRNA-182-3p and miRNA-155 possessed 11, 15 and 20 mismatched bases related to miRNA-182, respectively. In the presence of target miRNA-182, the SPR signal difference was 62 mDeg, suggesting the occurrence of the enzymatic cleavage reaction. However, in the cases of miRNA-96, miRNA-182-3p and miRNA-155, the hybridization between miRNA and blRNA was hindered to a larger extent, and the SPR signal differences of 17 mDeg, 12 mDeg and 4 mDeg were attained, respectively. The results demonstrated the capability of the method for distinguishing miRNA-182 from other similar sequences.
The dependence of SPR signals on the concentrations of miRNA-182 was investigated (Fig. 4, Fig. S3). The signal difference, defined as that in Fig. 3, was linearly proportional to miRNA concentrations ranging from 0.005 pM to 1.0 pM. The linear regression equation was expressed as Signal Difference (mDeg) = 24.20 [miRNA-182] (pM) + 10.08 (R2 = 0.985). The limit of detection (LOD) was estimated to be 0.62 fM based on 3 SD/Slope, where SD was the standard deviation of the blank measurements in the absence of miRNA-182. The relative standard deviations (RSDs) for three replicate measurements of all the miRNA concentrations were below 9%, indicating high reproducibility of the assay. The analytical performances of the proposed method were comparable with those of other signal-amplified SPR methods and DNA walking-based amplification methods (Table 2).
The feasibility of the proposed method for determination of the expression levels of miRNA-182 in serum samples from glioma patients and healthy donors has been demonstrated. The serum samples were diluted 5 times with PBS, and then treated by ultrafiltration membranes with a molecular weight cut-off of 10,000 Da to remove high-molecular-weight species. As shown in Fig. 5, the miRNA-182 was highly expressed in glioma patients, and the average expression levels of miRNA-182 from three glioma patients were 3.4 times higher than those from three healthy donors, being consistent with the results by electrochemistry and RT-PCR58,59. The SPR method based on DNA walking and enzyme cleavage for miRNA detection served as a promising alternative for point-of-care diagnosis of disease-related biomarkers.
Conclusion
In summary, SPR assay of miRNA based on DNA walking process and enzymatic cleavage reaction has been developed. The rationally designed protocol possessed high sensitivity and specificity due to the repeated hybridization and cleavage cycles occurring on the chip surface. The capability of the method for differentiating miRNA-182 with different concentrations and miRNAs with different sequences was demonstrated. The method exhibited a detection limit of 0.62 fM and was employed for quantificaiton of miRNA-182 in serum samples. The expression levels of miRNA-182 from glioma patients were 3.4 times higher than those from healthy donors, proving the reliability of the method for complicated samples analysis. The sensing strategy holds great potential for early diagnosis of major clinical diseases.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Esquela-Kerscher, A. & Slack, F. J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 (2006).
Kosaka, N., Iguchi, H. & Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 101, 2087–2092 (2010).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Liu, N. & Olson, E. N. MicroRNA regulatory networks in cardiovascular development. Dev. Cell 18, 510–525 (2010).
Junn, E. & Mouradian, M. M. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol. Ther. 133, 142–150 (2012).
Liang, R. Q. et al. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 33, e17 (2005).
Torres, A. G., Fabani, M. M., Vigorito, E. & Gait, M. J. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA 17, 933–943 (2011).
Chen, C. F. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005).
Schmieder, S. et al. Ultrasensitive SPR detection of miRNA-93 using antibody-enhanced and enzymatic signal amplification. Eng. Life Sci. 17, 1264–1270 (2017).
Hao, K. et al. High-sensitive surface plasmon resonance microRNA biosensor based on streptavidin functionalized gold nanorods-assisted signal amplification. Anal. Chim. Acta 954, 114–120 (2017).
Liu, R. et al. Surface plasmon resonance biosensor for sensitive detection of microRNA and cancer cell using multiple signal amplification strategy. Biosens. Bioelectron. 87, 433–438 (2017).
Wang, X. et al. In situ template generation of silver nanoparticles as amplification tags for ultrasensitive surface plasmon resonance biosensing of microRNA. Biosens. Bioelectron. 137, 82–87 (2019).
Zeng, K., Li, H. & Peng, Y. Gold nanoparticle enhanced surface plasmon resonance imaging of microRNA-155 using a functional nucleic acid-based amplification machine. Microchim. Acta 184, 2637–2644 (2017).
Zhang, D. et al. Streptavidin-enhanced surface plasmon resonance biosensor for highly sensitive and specific detection of microRNA. Microchim. Acta 180, 397–403 (2013).
Li, J. et al. An enzyme-free surface plasmon resonance biosensor for real-time detecting microRNA based on allosteric effect of mismatched catalytic hairpin assembly. Biosens. Bioelectron. 77, 435–441 (2016).
Li, X. et al. A novel surface plasmon resonance biosensor for enzyme-free and highly sensitive detection of microRNA based on multi component nucleic acid enzyme (MNAzyme)-mediated catalyzed hairpin assembly. Biosens. Bioelectron. 80, 98–104 (2016).
Wei, X. et al. An enzyme-free surface plasmon resonance imaging biosensing method for highly sensitive detection of microRNA based on catalytic hairpin assembly and spherical nucleic acid. Anal. Chim. Acta 1108, 21–27 (2020).
Aoki, H., Corn, R. M. & Matthews, B. MicroRNA detection on microsensor arrays by SPR imaging measurements with enzymatic signal enhancement. Biosens. Bioelectron. 142, 111565 (2019).
Wang, Q. et al. Surface plasmon resonance biosensor for enzyme-free amplified microRNA detection based on gold nanoparticles and DNA supersandwich. Sens. Actuators B 223, 613–620 (2016).
Wang, Q. et al. Graphene oxide-gold nanoparticles hybrids-based surface plasmon resonance for sensitive detection of microRNA. Biosens. Bioelectron. 77, 1001–1007 (2016).
Li, Q. et al. High sensitivity surface plasmon resonance biosensor for detection of microRNA and small molecule based on graphene oxide-gold nanoparticles composites. Talanta 174, 521–526 (2017).
Gu, Y. et al. Ultrasensitive microRNA assay via surface plasmon resonance responses of Au@Ag nanorods etching. Anal. Chem. 89, 10585–10591 (2017).
Ding, X. et al. Surface plasmon resonance biosensor for highly sensitive detection of microRNA based on DNA super-sandwich assemblies and streptavidin signal amplification. Anal. Chim. Acta 874, 59–65 (2015).
Dou, B., Zhou, H., Hong, Y., Zhao, L. & Wang, P. Cross-triggered and cascaded recycling amplification system for electrochemical detection of circulating microRNA in human serum. Chem. Commun. 57, 7116–7119 (2021).
Zhang, X. et al. Novel 2D DNA nanoprobe mediated enzyme-free target recycling amplification for the ultrasensitive electrochemical detection of microRNA. Anal. Chem. 90, 9538–9544 (2018).
Xu, S. et al. One DNA circle capture probe with multiple target recognition domains for simultaneous electrochemical detection of miRNA-21 and miRNA-155. Biosens. Bioelectron. 149, 111848 (2020).
Feng, Q.-M. et al. Ratiometric electrochemical detection of microRNA based on construction of a hierarchical C@SnS2 nanoflower sensing interface. Chin. J. Anal. Chem. 49, e21020–e21028 (2021).
Hong, C., Baek, A., Hah, S. S., Jung, W. & Kim, D.-E. Fluorometric detection of microRNA using isothermal gene amplification and graphene oxide. Anal. Chem. 88, 2999–3003 (2016).
Miao, X. et al. Label-free platform for microRNA detection based on the fluorescence quenching of positively charged gold nanoparticles to silver nanoclusters. Anal. Chem. 90, 1098–1103 (2018).
Wang, K. et al. Ultrasensitive detection of microRNA with isothermal amplification and a time-resolved fluorescence sensor. Biosens. Bioelectron. 57, 91–95 (2014).
Yuan, Y. et al. Highly-efficient luminol immobilization approach and exponential strand displacement reaction based electrochemiluminescent strategy for monitoring microRNA expression in cell. Biosens. Bioelectron. 132, 62–67 (2019).
Liu, Q. et al. A novel electrochemiluminescence biosensor for the detection of microRNAs based on a DNA functionalized nitrogen doped carbon quantum dots as signal enhancers. Biosens. Bioelectron. 92, 273–279 (2017).
Feng, X. et al. Ratiometric biosensor array for multiplexed detection of microRNAs based on electrochemiluminescence coupled with cyclic voltammetry. Biosens. Bioelectron. 75, 308–314 (2016).
Deng, H., Shen, W., Ren, Y. & Gao, Z. A highly sensitive and selective homogenous assay for profiling microRNA expression. Biosens. Bioelectron. 54, 650–655 (2014).
Piao, J. et al. Enzyme-free colorimetric detection of microRNA-21 using metal chelator as label for signal generation and amplification. Anal. Chim. Acta 1052, 145–152 (2019).
Xia, Y. et al. Colorimetric detection of exosomal microRNA through switching the visible-light-induced oxidase mimic activity of acridone derivate. Biosens. Bioelectron. 173, 112834 (2021).
Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 108, 462–493 (2008).
Jebelli, A., Oroojalian, F., Fathi, F., Mokhtarzadeh, A. & de la Guardia, M. Recent advances in surface plasmon resonance biosensors for microRNAs detection. Biosens. Bioelectron. 169, 112599 (2020).
Saha, K., Agasti, S. S., Kim, C., Li, X. & Rotello, V. M. Gold nanoparticles in chemical and biological sensing. Chem. Rev. 112, 2739–2779 (2012).
Dundas, C. M., Demonte, D. & Park, S. Streptavidin-biotin technology: Improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 97, 9343–9353 (2013).
Bi, S., Yue, S. & Zhang, S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 46, 4281–4298 (2017).
Yue, S., Li, Y., Qiao, Z., Song, W. & Bi, S. Rolling circle replication for biosensing, bioimaging, and biomedicine. Trends Biotechnol. 39, 1160–1172 (2021).
Wang, L. et al. Simple tripedal DNA walker prepared by target-triggered catalytic hairpin assembly for ultrasensitive electrochemiluminescence detection of microRNA. ACS Sens. 5, 3584–3590 (2020).
Hai, X. et al. DNA-based label-free electrochemical biosensors: From principles to applications. Trends Anal. Chem. 133, 116098 (2020).
Xu, M. & Tang, D. Recent advances in DNA walker machines and their applications coupled with signal amplification strategies: A critical review. Anal. Chim. Acta 1171, 338523 (2021).
Wang, D. et al. Highly selective detection of single-nucleotide polymorphisms using a quartz crystal microbalance biosensor based on the toehold-mediated strand displacement reaction. Anal. Chem. 84, 7008–7014 (2012).
Zhu, J., Ding, Y., Liu, X., Wang, L. & Jiang, W. Toehold-mediated strand displacement reaction triggered isothermal DNA amplification for highly sensitive and selective fluorescent detection of single-base mutation. Biosens. Bioelectron. 59, 276–281 (2014).
Lee, P. Y., Costumbrado, J., Hsu, C. Y. & Kim, Y. H. Agarose gel electrophoresis for the separation of DNA fragments. J. Vis. Exp. 62, e3923 (2012).
Antipova, V. N., Zheleznaya, L. A. & Zyrina, N. V. Ab initio DNA synthesis by Bst polymerase in the presence of nicking endonucleases Nt.AlwI, Nb.BbvCI, and Nb.BsmI. FEMS Microbiol. Lett. 357, 144–150 (2014).
Bellamy, S. R. W. et al. Cleavage of individual DNA strands by the different subunits of the heterodimeric restriction endonuclease BbvCI. J. Mol. Biol. 348, 641–653 (2005).
Heiter, D. F., Lunnen, K. D. & Wilson, G. G. Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI. J. Mol. Biol. 348, 631–640 (2005).
Wang, J. et al. Multiplexed electrochemical detection of miRNAs from sera of glioma patients at different stages via the novel conjugates of conducting magnetic microbeads and diblock oligonucleotide-modified gold nanoparticles. Anal. Chem. 89, 10834–10840 (2017).
He, L. et al. MicroRNA-182-3p negatively regulates cytokines expression by targeting TLR5M in orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol. 93, 589–596 (2019).
Li, M. et al. Sensitive electrochemical detection of microRNA based on DNA walkers and hyperbranched HCR-DNAzyme cascade signal amplification strategy. Sens. Actuators B 345, 130348 (2021).
Kuang, Q. et al. Three-way DNA junction structure combined with enzyme-powered cascade amplification for ultrasensitive electrochemiluminescence detection of microRNA via smart DNA walker. Sens. Actuators B 274, 116–122 (2018).
Li, Q. et al. Ratiometric fluorescent 3D DNA walker and catalyzed hairpin assembly for determination of microRNA. Microchim. Acta 187, 365 (2020).
Wang, L., Liu, Z.-J., Cao, H.-X. & Liang, G.-X. Ultrasensitive colorimetric miRNA detection based on magnetic 3D DNA walker and unmodified AuNPs. Sens. Actuators B 337, 129813 (2021).
Wang, J. et al. Direct quantification of microRNA at low picomolar level in sera of glioma patients using a competitive hybridization followed by amplified voltammetric detection. Anal. Chem. 84, 6400–6406 (2012).
Lu, H., Hailin, T., Yi, X. & Wang, J. Three-dimensional DNA nanomachine combined with toehold-mediated strand displacement reaction for sensitive electrochemical detection of miRNA. Langmuir 36, 10708–10714 (2020).
Acknowledgements
The authors thank the financial support of this work by the National Natural Science Foundation of China (Nos. 22076221, 21876208), the Innovation-Driven Project of Central South University (No. 2020CX005), the Hunan Provincial Science and Technology Plan Project, China (No. 2019TP1001), and the Program for Innovative Research Team of Science and Technology in the University of Henan Province (No. 21IRTSTHN005).
Author information
Authors and Affiliations
Contributions
J.W. and X.Y. wrote the main manuscript text and S.C. and Y.H. prepared all the Figures and the Tables. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, S., He, Y., Liu, L. et al. DNA walking system integrated with enzymatic cleavage reaction for sensitive surface plasmon resonance detection of miRNA. Sci Rep 12, 16093 (2022). https://doi.org/10.1038/s41598-022-20453-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20453-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.