Abstract
Globally, 94% of Treponema pallidum subsp. pallidum (TPA) clinical strains belong to the SS14-like group and 6% to the Nichols-like group, with a prevalence of macrolide resistance of 90%. Our goal was to determine whether local TPA strain distribution and macrolide resistance frequency have changed significantly since our last report, which revealed that Buenos Aires had a high frequency of Nichols-like strains (27%) and low levels of macrolide resistance (14%). Swab samples from patients with suspected syphilis were collected during 2015–2019 and loci TP0136, TP0548, TP0705 were sequenced in order to perform multilocus sequence typing. Strains were classified as Nichols-like or SS14-like. The presence of macrolide resistance-associated mutations was determined by examination of the 23S rDNA gene sequence. Of 46 typeable samples, 37% were classified as Nichols-like and 63% as SS14-like. Macrolide resistance prevalence was 45.7%. Seven allelic profiles were found, five were SS14-like and two were Nichols-like. The frequency of Nichols-like strains increased between studies (26.8% vs. 37%, p = 0.36). A dramatic increase was found in the frequency of macrolide resistant strains between studies (14.3% vs. 45.7%, p = 0.005). Our results are in agreement with international trends and underscore the need to pursue further TPA molecular typing studies in South America.
Similar content being viewed by others
Introduction
Treponema pallidum subsp. pallidum (TPA) is the bacterial pathogen that causes syphilis, a chronic venereal disease in humans1. Despite the availability of several diagnostic tools as well as the effective treatment with penicillin, syphilis prevalence and incidence remain high around the world. The World Health Organization estimated that approximately 22.3 million people globally between the ages of 15 and 49 had syphilis in 2020, with 7.1 million new syphilis cases per year globally, and 1.1 million new cases in the Region of the Americas2.
In Argentina, syphilis incidence has been steadily on the rise since 2010, with an increase from 21.2 new cases per 100,000 habitants in 2015 to 56.1 per 100,000 habitants in 2019, and 1.55 cases of congenital syphilis per 1000 live births in 20193. Several epidemiological studies have identified groups that are disproportionally vulnerable to syphilis, such as men who have sex with men, female sex workers and male to female transgender people, with syphilis prevalences between 17 and 50%4,5,6,7.
The molecular characterization of TPA has been of increasing interest in recent years as a useful tool for improving understanding of syphilis network transmission and epidemiology dynamics. Whole genome sequencing (WGS) of clinical strains from around the world reveals that the global syphilis population comprises just two deeply branching lineages, Nichols and SS14, each with multiple sublineages. The dominant TPA sublineages circulating today are believed to be the result of a rapid population expansion in the 2000s, following a population bottleneck in the 1990s8,9. The first typing scheme was designed by the Center for Disease Control and Prevention (CDC) in 199810 and subsequently improved11,12. An alternative sequencing-based molecular typing (SBMT) scheme was introduced in 200613 and recently enhanced14. The latest multilocus sequence typing (MLST) scheme analyzes the TP0136, TP0548 and TP0705 loci and provides a simple alternative to WGS, retaining around 30% of WGS’s resolution power. Additionally, MLST allows for the distinction between the two TPA clades, SS14 and Nichols (based on TP0136 and TP0548 sequences), as well as the differentiation of strains within each clade (based on TP0136, TP0548 and TP0705 sequences). MLST also has the advantage of distinguishing TPA from other treponemes such as Treponema pallidum subsp. pertenue and Treponema pallidum subsp. endemicum14. MLST of TPA strains has been expanding since its proposal: the TPA PubMLST database15 currently contains 1572 strain records from 40 different countries. However, over 70% of records come from Europe. Both SBMT and MLST can be paired with the analysis of 23S rDNA to determine the presence of mutations associated with macrolide resistance.
We have previously conducted the first characterization of circulating TPA strains in Buenos Aires, Argentina using SBMT, in samples collected between 2006 and 2013. We detected the presence of nine distinct allelic profiles, five of which had not been described before. Out of these nine haplotypes, 77% were related to SS14 and 23% to Nichols strains16. These results suggested that the syphilis epidemic in Argentina differed considerably from the global distribution pattern, according to which Nichols-like strains are much rarer17.
Even though penicillin is the antibiotic of choice for the treatment of syphilis, there are other antibiotics which can be prescribed in certain situations (e.g. penicillin allergy). Macrolides in particular are a desirable treatment option because of their oral administration, which makes them more readily acceptable by patients. Two point mutations in the 23S rDNA locus (A2058G and A2059G) have been linked to macrolide resistance in TPA strains18. Global prevalence of macrolide resistance mutations is 90%, with wide regional variations: Cuba, the US and several European countries report high prevalence rates (61–100%), whereas Canada, Russia, South Africa, Madagascar, Taiwan and Peru report little to no macrolide resistance (0–12%). Our first study seemed to indicate that the situation in Argentina was closer to the latter, with 14% prevalence of macrolide resistance mutations16.
Several studies have shown that both the predominant TPA strains and the frequency of mutations associated with macrolide resistance in a particular population can change over time12,19,20. With the aim of studying the dynamics of TPA strains in Buenos Aires, Argentina, the purpose of the present study was to characterize the syphilis epidemic in Buenos Aires by analyzing TPA clinical strains collected during 2015–2019 in terms of their strain and haplotype distribution as well as the presence of antibiotic resistance-associated mutations, in order to update and expand on our previous report using the newer MLST methodology.
Results
Characteristics of patients and clinical samples
A total of 99 swabs were collected from patients with suspected syphilis lesions (Fig. 1). In 28 cases, active syphilis was ruled out due to negative serology (non reactive Venereal Disease Research Laboratory –VDRL– test), which was also confirmed by negative PCR in all cases. Among syphilis-positive patients, DNA isolation was unsuccessful in 14 cases, as evidenced by negative beta-actin PCR. Treponemal DNA (polA and/or tmpC) amplification was unsuccessful in 11 cases.
Clinical characteristics of patients with syphilis diagnosis (based on serology and dark-field microscopy –DFM– results) and successful DNA isolation are presented in Table 1. Briefly, 83% of patients were men with a median age of 25 years and 46% of those with known HIV status were HIV-positive. Most patients were Argentine nationals (74%) and unmarried (88%), with approximately half of them residing in the city of Buenos Aires (52%) or the Province of Buenos Aires (48%); 52% were students or unemployed. Lesions were mainly located in the genitals (53%), some were oral (28%) and only one was an anal lesion. Most patients (83%) had positive VDRL serology, 65% with high titres (> 1:32). Although data on syphilis stage could not be recovered for almost half of the patients, among those with a determined clinical stage, 45% were primary syphilis cases and 55% were secondary syphilis cases. No significant differences were found between patients with PCR-positive and PCR-negative samples in terms of sociodemographic or clinical characteristics.
Multilocus sequence typing (MLST)
All 46 PCR-positive samples were fully or partially typeable using MLST. Samples were considered typeable when at least one sequence of a typing locus (either TP0136, TP0548 or TP0705) was obtained. By comparing the TP0136 and/or TP0548 sequences from our samples with reference sequences we determined that 63% (29/46) of strains belonged to the SS14-like group and 37% (17/46) to the Nichols-like group. In all cases where both TP0136 and TP0548 were successfully sequenced (n = 37), clade assignment was consistent between loci.
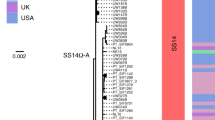
Samples from 34 (73.9%) patients were completely typed (including the TP0136, TP0548 and TP0705 gene sequences), 29 also including the 23S rDNA gene sequence. Altogether, seven different TPA haplotypes were found among the fully typed: five were related to the sequence from the TPA strain SS14 (1.1.1, 1.3.1, 1.1.8, 1.32.1 and 1.48.3) and two were related to the sequence from the TPA strain Nichols (9.7.3 and 9.20.3). All identified haplotypes and their frequencies are shown in Table 2. Briefly, the most frequent allelic profile was 9.7.3 (35.3% among fully typed samples) followed by 1.1.1 and 1.3.1 (each 26.5% among fully typed samples), and finally by 1.1.8, 1.32.1, 1.48.3 and 9.20.3 with only one case each. Among the partially typed samples, only one SS14-like strain showed an haplotype that was not consistent with any of the fully typed profiles (X.1.3) All alleles found in this study have been described previously; however, we describe the new allelic profile 1.48.3. A phylogenetic analysis of allelic profiles found among fully typed samples is shown in Fig. 2.
Majority rule consensus tree of 1000 bootstrap replicates performed on concatenated TP0136, TP0548 and TP0705 sequences identified among fully typed samples. Bootstrap percentages given for clades recovered with more than 50% support. The scale shows the number of substitutions per site. Bootstrap values are shown next to branches. The length of concatenated sequences was 1901 bp and contained 44 variable positions. The tree was constructed in MEGA X52 using the Maximum Likelihood method54 with the bootstrap test55.
Prevalence of mutations associated with macrolide resistance
The 23S rDNA gene locus was amplified in 35 samples, and in 16 of them (45.7%), either the A2058G (15 patients, 93.7%) or the A2059G (1 patient, 6.3%) mutation was found. No strains harbored both A2058G and A2059G mutations. The sequences of both copies of the 23S rRNA gene were identical in all cases. Macrolide resistance was detected in both SS14-like (13/22) and Nichols-like (3/13) strains. The frequency of resistance was higher in the SS14 clade, however without statistical significance (59.1% vs. 23.1%, p = 0.08).
Associations of clade and allelic profiles with patient characteristics
We found no statistically significant associations of clade or allelic profile with patient age, gender, residence, nationality, occupation, marital status, sexually transmitted infection (STI) history, location of lesion, VDRL titer, DFM results, syphilis stage or HIV status. Patients carrying Nichols-like strains tended to be slightly older than patients carrying SS14-like strains (median age: 31 vs. 25 years, p = 0.07) and to have higher VDRL titers (median titer: 128 vs. 32, p = 0.09).
Discussion
At the local level, this study is the second to analyze the genetic characteristics of circulating TPA in Argentina, serving as both a continuation and an update of the previous report by our research group16. While the first study used the SBMT methodology, this study is the first to apply the enhanced MLST scheme in Argentina. At the regional level, this study constitutes as well one of the first initiatives to characterize the diversity of TPA in South America, where only a few molecular epidemiology studies of TPA have been conducted21,22, and even fewer using the MLST methodology23.
In this study, as in most reports worldwide, the SS14 clade was predominant over the Nichols clade. However, the frequency of Nichols-like strains observed (37%) is considerably higher than is usually the case globally. Clade SS14 is reported to constitute the vast majority (94.1%) of worldwide TPA clinical strains, though the relative frequencies of these clades show some geographical variation: while in most countries the prevalence of Nichols-like strains is lower than 10%, this has been found to be 15% in Japan24, 21.4% in Peru, 20.2% in Taiwan and 100% in Madagascar25. In Argentina, our first study16 showed a frequency of Nichols-like strains of 26.8%, which would make Argentina the country with the highest reported frequency at the time after Madagascar. In the present study we found an even higher prevalence of the Nichols clade which might indicate that this prevalence is currently on the rise. Moreover, our results, together with those of the report from Peru22, suggest that South America is a region with a high frequency of Nichols-like circulating strains, though admittedly more data is required to support this claim. However, it must be noted that, although the city of Buenos Aires and its surroundings account for a considerable percentage of the Argentine population, our results may not reflect TPA diversity in other parts of the country.
The explanation for the higher frequency of Nichols-like strains observed in some countries remains elusive, considering that genomic comparisons of these strains do not reveal differences in genes known to confer a fitness advantage8. A higher frequency of macrolide resistance in SS14-like strains, like we observed in this study, might explain its overall predominance, but their expansion due to other selective advantages cannot be excluded.
Among fully typed samples, we found seven distinct haplotypes, five of which were related to the SS14 strain (1.1.1, 1.3.1, 1.1.8, 1.32.1 and 1.48.3), while the other two were related to the Nichols strain (9.7.3 and 9.20.3). Haplotypes 9.7.3 (35%), 1.1.1 (27%) and 1.3.1 (27%) accounted for most samples. In our previous study, a different SBMT nomenclature was used and also that study did not include the TP0705 locus, making comparisons difficult. However, a rough comparison clearly reveals that the most common haplotypes have changed. In our previous study, the SS14-like strains SSS, SSR9 and SSR8 (now named 1.1.X) constituted the most prevalent (43.9%), followed by the Nichols-like strains U3U6S and U3U6R8 (now named 9.7.X) representing 17% of the cases. However, in this new study, strain 9.7.X (specifically 9.7.3) constitutes the most prevalent (35.3%) followed by strain 1.1.X (29.4%, including 1.1.1., 26.5%, and 1.1.8, 2.9%) and strain 1.3.X (specifically 1.3.1., 26.5%). Thus, although SS14-like strains as a whole remained dominant, the most prevalent allelic profile changed from an SS14-related haplotype to a Nichols-related haplotype, reflecting the increase in the overall frequency of Nichols strains. The difference in haplotype distribution between the Argentinean studies may be due to the fact that samples were collected at different times (2006–2013 vs. 2015–2019); several studies show that TPA strains in a particular population can vary over time12,19,20. Although both studies were aimed at the same population, recruited at the same clinic, differences in TPA diversity may also be partially explained by variations in demographic and clinical characteristics: compared to the previous study, the results presented here correspond to a sample that includes more women (female percentage: 19.6% vs. 4.7%, p = 0.03) and less primary syphilis cases (percentage of primary cases among those with determined stage: 46.2% vs. 73.7%, p = 0.03).
A comparison of the frequencies of TPA allelic profiles observed in this study with the average frequencies reported in other continents (Table 3) shows that, not only are the relative frequencies of the SS14 and Nichols clades observed in Argentina unusual, but so are the allelic profiles detected. Haplotype 9.7.3, which was found in 35% of our samples, is only present in less than 5% of samples in other continents. Meanwhile haplotype 1.3.1, which accounts for roughly half of clinical samples in Europe and 60% of samples in North America, was found in approximately a fourth of Argentinian samples. Haplotype 1.1.1, though not rare in other continents, was detected in our samples with a considerably higher frequency than usually reported15,26. These differences underscore the need for further TPA molecular typing studies in South America, since it cannot be assumed that clade and allelic profile frequencies among local clinical samples will be similar to those in other regions.
Considering that the nucleotide diversity has been demonstrated to be higher within the strains belonging to the Nichols clade than within the strains belonging to the SS14 clade (π = 0.05 for Nichols and π = 0.01 for SS1417), and given the higher frequency of Nichols-like strains in our samples, one might expect a higher number of different Nichols-like haplotypes than were found in this study. It must be noted however, that only a limited number of positive samples (46) and detected haplotypes (seven) were obtained, thus it is possible that a lower number of Nichols haplotypes was detected by chance. At the same time, it has been reported that the frequency of some genotypes expands or decreases over time19,27.
The observed prevalence of macrolide resistance-associated mutations in the present study was nearly 50%, the A2058G mutation accounting for all but one of these cases. The presence of this mutation was detected in SS14-like as well as Nichols-like strains, with a tendency towards higher mutation frequency among SS14-like strains (59% vs. 23%), which was also the case in our first study16. The frequency of macrolide resistance has tripled between our first study and this study (14.3% vs 45.7%, p < 0.005) and it places Argentina closer to other countries with high prevalence of macrolide resistance such as Australia28, Cuba27, Czech Republic19,23, China29,30,31,32, Ireland33,34, Spain35, the UK36 and the US11,37,38,39. As shown in Table 4, even though the presence of macrolide resistance mutations increased with time in both clades, this increase showed statistical significance only in the SS14 clade. The observed increase in the frequency of macrolide-resistant TPA strains in our study population coincides with the global trend40.
This rise might reflect a change in the consumption of macrolide antibiotics in Argentina. However, data on antibiotic use in our country is limited: a study on the evolution of the purchase of antibiotic drugs from Argentine pharmacies between 2015 and 2017 found no substantial change in the purchase of macrolides41. Another explanation could be an increased veterinary use of macrolides, as is the case with other pathogens42, and/or increased travelling. More publicly available information on medical and veterinary uses of macrolides in Argentina would be necessary to shed light on the observed increase in macrolide resistance-associated mutations in circulating TPA strains.
Of the 99 swab samples collected from patients with clinical suspicion of syphilis, 28 were ruled out as syphilis-negative. Among the remaining 71 samples, DNA isolation was unsuccessful in 14 cases, while treponemal DNA amplification was unsuccessful in 11 cases. These failures in DNA isolation and amplification may be due to low amounts of material collected during swabbing, to the presence of PCR inhibitors or to DNA degradation between swab collection and storage at − 20 °C. Since the exact amount of time elapsed was not recorded for all samples, it is not possible to verify whether there is a correlation between time at room temperature and amplification failure.
In this study, no statistically significant associations were found between clade or allelic profile and patient characteristics. However, patients carrying Nichols-like strains tended to be slightly older than patients carrying SS14-like strains. A significant association between the Nichols clade and older patients has been reported in clinical samples from France43. The Nichols clade also tended to be associated with higher VDRL titers, but this association has not, to our knowledge, been found in other studies.
Conclusions
Molecular typing studies of human pathogenic bacteria can help elucidate the evolutionary history and origin of old diseases such as syphilis17, while also contributing to the understanding of transmission mechanisms, epidemic diversity and dynamics, as well as to the improvement of clinical practice and public policies of control and prevention44. Particularly in the case of TPA, a bacterium which has until recently45 resisted any attempt at long-term in vitro culture, molecular typing techniques prove especially useful. Global TPA typing data has facilitated the characterization of syphilis outbreaks, the discovery of associations between subtypes and neurosyphilis, the surveillance of macrolide resistance, the distinction between re-infection and reactivation, and the comprehension of the geographical, temporal and population distribution of TPA.
However, in spite of all its benefits, only a limited number of studies from a few countries have focused on molecular typing of TPA since the emergence of the first typing assays, especially in South America44. This study attempts to fill this gap of knowledge. Our results suggests that the prevalence of circulating TPA strains in our region may differ considerably from those reported in other regions, while also changing over time. Although the SS14 clade was shown to be dominant, the prevalence of the Nichols clade was higher than in most countries, and higher than in our previous study. Frequencies of allelic profiles also differed considerably from those reported in other regions and in our previous report. Notably, the prevalence of macrolide resistance-associated mutations increased dramatically between studies; the causes of this rise are of importance to public health and warrant further investigation. In addition to this, future molecular epidemiology studies of syphilis should aim at providing useful information for the development of prevention and control programs, focusing for instance on the identification of at-risk populations and sources of infection, as well as the verification of links between certain subtypes and pathology.
Methods
Clinical samples
Clinical swab samples were collected between May 2015 and November 2019 from patients with suspected primary or secondary syphilis at an STI clinic in the city of Buenos Aires (Programa de Enfermedades de Transmisión Sexual –PETS-, Hospital de Clínicas “José de San Martín”, University of Buenos Aires, Argentina). Although it is possible to detect treponemal DNA in non-lesion samples, lesions tend to have a higher treponemal load and thus usually yield higher DNA amplification efficiencies, therefore swab samples were taken only from lesions46. Swab samples from genital, anal, oral or rectal lesions were taken by a trained dermatologist, without cleaning the lesion. Samples were handled as described previously47,48,49. The swab samples remained at room temperature for no more than a week before being stored at − 20 °C until DNA isolation.
Patients suspected of having syphilis based on clinical findings were diagnosed with laboratory tests that included the examination of lesion material by DFM and serological tests. Serology included VDRL, FTA-ABS and TPHA50. Patients were considered to have syphilis when one of the tests (DFM and/or serology) was positive. Patients gave their written, informed consent and received recommendations of treatment, care, referral to the laboratory for serological diagnosis, and a new visit for follow-up. Clinical data included patients’ age, gender, STI history, location of lesion, results of serology and DFM, syphilis stage and HIV status. Socio-demographic data included residence, nationality, occupation and marital status.
DNA isolation
DNA isolation from swabs was performed using a commercial kit (QIAamp DNA Blood Mini Kit, Qiagen, Germany). First, swabs were submerged in 600 µl of phosphate buffered saline. The eluate was then processed according to the manufacturer’s recommendations. The efficacy of the extraction process was verified by PCR amplification of the human beta-actin gene. The PCR mixture (final volume of 25 μl) contained 16.3 μl of water, 0.5 μl of a 10 mM deoxynucleotide triphosphate (dNTP) mixture, 2.5 μl of 10 × buffer, 3 μl of 25 mM MgCl2, 0.75 μl of each primer (10 μM), 0.25 μl of Taq DNA polymerase (5000 U/ml), and 1 μl of isolated total DNA. PCR amplification was performed under the following conditions: 95 °C (2 min); 94 °C (30 s), 60 °C (30 s), 72 °C (30 s) for 35 cycles; and 72 °C (7 min). Primers used can be found in Supplementary Table S1 online.
Detection of treponemal DNA
In order to screen for TPA-positive samples, amplification of loci TP0105 (polA) and TP0319 (tmpC) was performed by nested PCR, as described previously20. The PCR mixture (final volume of 25 μl) contained in the first step 15.3 μl of water, 0.1 μl of a 10 mM dNTP mixture, 2.5 μl of 10 × buffer, 1.5 μl of 25 mM MgCl2, 0.25 μl of each primer (10 μM), 0.1 μl of Taq DNA polymerase (5000 U/ml), and 5 μl of isolated DNA. PCR amplification was performed under the following conditions: 94 °C (1 min); 94 °C (30 s), 58 °C (30 s), 72 °C (1 min) for 30 cycles; and 72 °C (10 min). The mixture for the second step was the same except that it contained 19.3 μl of water and 1 μl of PCR product from the first step. PCR amplification was performed under the following conditions: 94 °C (1 min); 94 °C (30 s), 58 °C (30 s), 72 °C (1 min) for 40 cycles; and 72 °C (10 min). Samples were considered PCR-positive for treponemal DNA when PCR amplification was positive for at least one treponemal locus. Primers used can be found in Supplementary Table S1 online.
PCR amplification for MLST
MLST was performed to type all TPA-positive samples as described previously14,15,43,51. Four loci, including TP0136, TP0548, TP0705 and 23S rDNA genes, were amplified using the nested PCR protocol (both copies of the 23S rDNA locus were amplified). The PCR mixture (final volume of 25 μl) contained in both steps 20.5 μl of water, 0.5 μl of a 10 mM dNTP mixture, 2.5 μl of 10 × buffer, 0.25 μl of each primer (100 μM), 0.05 μl of Taq DNA polymerase (5000 U/ml), and 1 μl of isolated DNA. In the first step, PCR amplification was performed under the following conditions: 95 °C (1 min); 94 °C (30 s), 50 °C (30 s), 72 °C (1 min, 45 s) for 40 cycles; and 72 °C (7 min). In the second step, PCR amplification was performed under the following conditions: 95 °C (1 min); 94 °C (30 s), 55 °C (30 s), 72 °C (1 min, 45 s) for 40 cycles; and 72 °C (7 min). The second amplification step was repeated an additional three times, in order to quadruple the volume of PCR product per sample prior to DNA purification. PCR products were purified with polyethylene glycol (PEG), using an in-house protocol (see Supplementary Information). Primers used can be found in Supplementary Table S1 online.
DNA sequencing and sequence analysis
Sequencing reactions were performed on the purified PCR products using a commercial kit (BigDye™ Terminator v3.1 Cycle Sequencing Kit, Applied Biosystems, USA). The products were sequenced on an automated capillary DNA sequencing system. Sequence analyses were performed using Sequencher software (Sequencher® Version 5.4.6 DNA Sequence Analysis Software, Gene Codes Corporation, Ann Arbor, MI, USA). TP0136 and TP0548 sequences were aligned to reference sequences TPA Nichols (CP004010.2) and TPA SS14 (CP004011.1) in order to assign samples to the corresponding clade (clade assignment was possible where at least one of these loci was successfully sequenced). 23S rDNA sequences were evaluated at positions 2058 and 2059 in the 23S rDNA gene of Escherichia coli (accession no. V00331), where A → G mutations are associated with macrolide resistance. Alleles encoding resistance were marked A2058G or A2059G depending on the site of substitution. Both copies of the rDNA gene were sequenced and analysed.
Phylogenetic analyses
Clade assignment was confirmed through phylogenetic analysis: the software MEGA X52 was used to construct multiple sequence alignments for each gene separately via the ClustalW algorithm53, as well as for a concatenation of TP0136, TP0548 and TP0705 sequences, which were then used for the construction of phylogenetic trees, using the maximum likelihood method54 with bootstrapping (1000 replications, cutoff value of 50%)55 and the Tamura-Nei model. MLST of TPA is based on the different allelic profiles (haplotypes) described by a 3-letter code, where the first number corresponds to the TP0136 allele, the second to the TP0548 allele, and the third to the TP0705 allele (e.g. 1.3.1).
Statistical analyses
Comparisons were performed using Fisher's exact test, for categorical variables, and the Mann–Whitney U test, for numerical variables, via the SPSS software (IBM Corp. Released 2011. IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp).
Ethics statement
This study was approved by the Ethics Committee of the Hospital de Clínicas “José de San Martín” (University of Buenos Aires). All patients gave their written, informed consent. The study was conducted according to the principles expressed in the Declaration of Helsinki56, as well as national regulations regarding research.
Data availability
All alleles found in this study were identical to previously reported sequences, which can be found in the PubMLST database15.
References
Radolf, J. D. et al. Treponema pallidum, the syphilis spirochete: Making a living as a stealth pathogen. Nat. Rev. Microbiol. 14, 744–759 (2016).
World Health Organization. Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021: Accountability for the global health sector strategies 2016–2021: Actions for impact: Web annex 2: Data methods. https://apps.who.int/iris/bitstream/handle/10665/342813/9789240030992-eng.pdf?sequence=1&isAllowed=y (2021).
Dirección de Sida y ETS & Ministerio de Salud de la Nación. Boletín N° 38: Respuesta al VIH y las ITS en la Argentina. https://bancos.salud.gob.ar/recurso/boletin-ndeg-38-respuesta-al-vih-y-las-its-en-la-argentina (2021).
Pando, M. A. et al. Prevalence of HIV and other sexually transmitted infections among female commercial sex workers in Argentina. Am. J. Trop. Med. Hyg. 74, 233–238 (2006).
Pando, M. A. et al. HIV and other sexually transmitted infections among men who have sex with men recruited by RDS in Buenos Aires, Argentina: High HIV and HPV infection. PLoS ONE 7, e39834 (2012).
Pando, M. A. et al. Violence as a barrier for HIV prevention among female sex workers in Argentina. PLoS ONE 8, e54147 (2013).
dos Ramos Farías, M. S. et al. First report on sexually transmitted infections among trans (male to female transvestites, transsexuals, or transgender) and male sex workers in Argentina: High HIV, HPV, HBV, and syphilis prevalence. Int. J. Infect. Dis. 15, e635–e640 (2011).
Beale, M. A. et al. Global phylogeny of Treponema pallidum lineages reveals recent expansion and spread of contemporary syphilis. Nat. Microbiol. 6, 1549–1560 (2021).
Pla-Díaz, M. et al. Evolutionary processes in the emergence and recent spread of the syphilis agent, Treponema pallidum. Mol. Biol. Evol. 39, 318 (2022).
Pillay, A. et al. Molecular subtyping of Treponema pallidum subspecies pallidum. Sex. Transm. Dis. 25, 408–414 (1998).
Katz, K. A. et al. Molecular epidemiology of syphilis—San Francisco, 2004–2007. Sex. Transm. Dis. 37, 660–663 (2010).
Marra, C. M. et al. Enhanced molecular typing of Treponema pallidum: Geographical distribution of strain types and association with neurosyphilis. J. Infect. Dis. 202, 1380–1388 (2010).
Flasarová, M. et al. Molekulární detekce a typizace Treponema pallidum subsp. pallidum v klinickém materiálu. Epidemiol. Mikrobiol. Imunol. 55, 105–111 (2006).
Grillová, L. et al. Molecular characterization of Treponema pallidum subsp. pallidum in Switzerland and France with a new multilocus sequence typing scheme. PLoS ONE 13, e0200773 (2018).
Grillová, L., Jolley, K., Šmajs, D. & Picardeau, M. A public database for the new MLST scheme for Treponema pallidum subsp. pallidum: Surveillance and epidemiology of the causative agent of syphilis. PeerJ 6, e6182 (2019).
Gallo Vaulet, L. et al. Molecular typing of Treponema pallidum isolates from Buenos Aires, Argentina: Frequent Nichols-like isolates and low levels of macrolide resistance. PLoS ONE 12, e0172905 (2017).
Arora, N. et al. Origin of modern syphilis and emergence of a pandemic Treponema pallidum cluster. Nat. Microbiol. 2, 16245 (2017).
Stamm, L. V. Syphilis: Antibiotic treatment and resistance. Epidemiol. Infect. 143, 1567–1574 (2015).
Grillová, L. et al. Molecular typing of Treponema pallidum in the Czech Republic during 2011 to 2013: Increased prevalence of identified genotypes and of isolates with macrolide resistance. J. Clin. Microbiol. 52, 3693–3700 (2014).
Flasarová, M. et al. Sequencing-based molecular typing of Treponema pallidum strains in the Czech Republic: All identified genotypes are related to the sequence of the SS14 strain. Acta Derm. Venerol. 92, 669–674 (2012).
Cruz, A. R. et al. Secondary syphilis in Cali, Colombia: New concepts in disease pathogenesis. PLoS Negl. Trop. Dis. 4, e690 (2010).
Flores, J. A. et al. Treponema pallidum pallidum genotypes and macrolide resistance status in syphilitic lesions among patients at 2 sexually transmitted infection clinics in Lima, Peru. Sex. Transm. Dis. 43, 465–466 (2016).
Lieberman, N. A. P. et al. Treponema pallidum genome sequencing from six continents reveals variability in vaccine candidate genes and dominance of Nichols clade strains in Madagascar. PLoS Negl. Trop. Dis. 15, e0010063 (2021).
Nishiki, S., Lee, K., Kanai, M., Nakayama, S. I. & Ohnishi, M. Phylogenetic and genetic characterization of Treponema pallidum strains from syphilis patients in Japan by whole-genome sequence analysis from global perspectives. Sci. Rep. 11, 3154 (2021).
Šmajs, D., Strouhal, M. & Knauf, S. Genetics of human and animal uncultivable treponemal pathogens. Infect. Genet. Evol. 61, 92–107 (2018).
Fernández-Naval, C. et al. Multilocus sequence typing of Treponema pallidum subsp. pallidum in Barcelona. Future Microbiol. 16, 967–976 (2021).
Noda, A. A., Matos, N., Blanco, O., Rodríguez, I. & Stamm, L. V. First report of the 23S rRNA gene A2058G point mutation associated with macrolide resistance in Treponema pallidum from syphilis patients in Cuba. Sex. Transm. Dis. 43, 332–334 (2016).
Read, P. et al. Treponema pallidum strain types and association with macrolide resistance in Sydney, Australia: New TP0548 gene types identified. J. Clin. Microbiol. 54, 2172–2174 (2016).
Xiao, Y. et al. Molecular subtyping and surveillance of resistance genes in Treponema pallidum DNA from patients with secondary and latent syphilis in Hunan, China. Sex. Transm. Dis. 43, 310–316 (2016).
Martin, I. E., Gu, W., Yang, Y. & Tsang, R. S. W. Macrolide resistance and molecular types of Treponema pallidum causing primary syphilis in Shanghai, China. Clin. Infect. Dis. 49, 515–521 (2009).
Li, Z. et al. Two Mutations associated with macrolide resistance in Treponema pallidum in Shandong, China. J. Clin. Microbiol. 51, 4270–4271 (2013).
Chen, X.-S. et al. High prevalence of azithromycin resistance to Treponema pallidum in geographically different areas in China. Clin. Microbiol. Infect. 19, 975–979 (2013).
Lukehart, S. A. et al. Macrolide resistance in Treponema pallidum in the United States and Ireland. N. Engl. J. Med. 351, 154–158 (2004).
Muldoon, E. G., Walsh, A., Crowley, B. & Mulcahy, F. Treponema pallidum azithromycin resistance in Dublin, Ireland. Sex. Transm. Dis. 39, 784–786 (2012).
Fernández-Naval, C. et al. Enhanced molecular typing and macrolide and tetracycline-resistance mutations of Treponema pallidum in Barcelona. Future Microbiol. 14, 1099–1108 (2019).
Tipple, C., McClure, M. O. & Taylor, G. P. High prevalence of macrolide resistant Treponema pallidum strains in a London centre. Sex. Transm. Infect. 87, 486–488 (2011).
Grimes, M. et al. Two mutations associated with macrolide resistance in Treponema pallidum: Increasing prevalence and correlation with molecular strain type in Seattle, Washington. Sex. Transm. Dis. 39, 954–958 (2012).
Marra, C. M. et al. Antibiotic selection may contribute to increases in macrolide-resistant Treponema pallidum. J. Infect. Dis. 194, 1771–1773 (2006).
Su, J. R. Prevalence of the 23S rRNA A2058G point mutation and molecular subtypes in Treponema pallidum in the United States, 2007 to 2009. Sex. Transm. Dis. 39, 794–798 (2012).
Šmajs, D., Grillová, L. & Paštěková, L. Macrolide resistance in the syphilis spirochete, Treponema pallidum ssp. pallidum: Can we also expect macrolide-resistant yaws strains?. Am. J. Trop. Med. Hyg. 93, 678–683 (2015).
Maito, M. A. Use of commercialized antibiotics in Argentine Republic pharmacies (2015–2017). Revista Ciencia Reguladora 3, 30–34 (2018).
Kenyon, C. Positive association between the use of macrolides in food-producing animals and pneumococcal macrolide resistance: A global ecological analysis. Int. J. Infect. Dis. 116, 344–347 (2022).
Pospíšilová, P. et al. Multi-locus sequence typing of Treponema pallidum subsp. pallidum present in clinical samples from France: Infecting treponemes are genetically diverse and belong to 18 allelic profiles. PLoS ONE 13, e0201068 (2018).
Peng, R.-R. et al. Molecular typing of Treponema pallidum: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 5, e1273 (2011).
Edmondson, D. G., Hu, B. & Norris, S. J. Long-term in vitro culture of the syphilis spirochete Treponema pallidum subsp. pallidum. MBio 9, e01153-18 (2018).
Towns, J. M. et al. Treponema pallidum detection in lesion and non-lesion sites in men who have sex with men with early syphilis: A prospective, cross-sectional study. Lancet Infect. Dis. 21, 1324–1331 (2021).
Heymans, R. et al. Clinical value of Treponema pallidum real-time PCR for diagnosis of syphilis. J. Clin. Microbiol. 48, 497–502 (2010).
Shields, M., Guy, R. J., Jeoffreys, N. J., Finlayson, R. J. & Donovan, B. A longitudinal evaluation of Treponema pallidum PCR testing in early syphilis. BMC Infect. Dis. 12, 353 (2012).
World Health Organization. Sífilis. In Diagnóstico de laboratorio de las infecciones de transmisión sexual, incluida la infección por el virus de la inmunodeficiencia humana (eds Unemo, M. et al.) 115–139 (World Health Organization, 2014).
Peeling, R. W. et al. Syphilis. Nat. Rev. Dis. Primers 3, 17073 (2017).
Vrbová, E. et al. MLST typing of Treponema pallidum subsp. pallidum in the Czech Republic during 2004–2017: Clinical isolates belonged to 25 allelic profiles and harbored 8 novel allelic variants. PLoS ONE 14, e0217611 (2019).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Tamura, K., Nei, M. & Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035 (2004).
Felsenstein, J. Confidence limits on phylogenies: An Approach using the bootstrap. Evolution 39, 783–791 (1985).
World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 310, 2191–2194 (2013).
Acknowledgements
We would like to thank all participants who made this study possible. This work was supported through a grant provided by the Universidad de Buenos Aires (UBACYT 20720130200010BA) awarded to Pando MA. The funding source had no role in the design or carrying out of the study, data collection, analysis, or interpretation, the writing of the manuscript, or the decision to submit it for publication.
Author information
Authors and Affiliations
Contributions
D.S. and M.A.P. conceived the study. A.M. enrolled participants. N.M. performed experiments. N.M., E.V., D.S., R.D.R. and M.A.P. analysed the data. N.M. and M.A.P. drafted the report. All authors read the manuscript, provided input into the report and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Morando, N., Vrbová, E., Melgar, A. et al. High frequency of Nichols-like strains and increased levels of macrolide resistance in Treponema pallidum in clinical samples from Buenos Aires, Argentina. Sci Rep 12, 16339 (2022). https://doi.org/10.1038/s41598-022-20410-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20410-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.