Abstract
Induced pluripotent stem cells (iPSCs) are a valuable resource for neurological disease-modeling and drug discovery due to their ability to differentiate into neurons reflecting the genetics of the patient from which they are derived. iPSC-derived cultures, however, are highly variable due to heterogeneity in culture conditions. We investigated the effect of passage number on iPSC differentiation to optimize the generation of sensory neurons (iPSC-dSNs). Three iPSC lines reprogrammed from the peripheral blood of three donors were differentiated into iPSC-dSNs at passage numbers within each of the following ranges: low (5–10), intermediate (20–26), and high (30–38). Morphology and pluripotency of the parent iPSCs were assessed prior to differentiation. iPSC-dSNs were evaluated based on electrophysiological properties and expression of key neuronal markers. All iPSC lines displayed similar morphology and were similarly pluripotent across passage numbers. However, the expression levels of neuronal markers and sodium channel function analyses indicated that iPSC-dSNs differentiated from low passage numbers better recapitulated the sensory neuron phenotype than those differentiated from intermediate or high passage numbers. Our results demonstrate that lower passage numbers may be better suited for differentiation into peripheral sensory neurons.
Similar content being viewed by others
Introduction
Induced pluripotent stem cells (iPSCs) are a valuable resource for neurological disease-modeling and drug discovery, largely due to their relative accessibility and capacity to differentiate into neurons with the same genetic background as the patient from which they are derived1,2,3,4,5,6. Studies on neurological disease usually have limited accessibility to human nerve tissue due to the potential for significant pain and permanent nerve damage during biopsy. Thus far, while animal models and immortalized primary neuron culture models have been used, there remain significant translational limitations. Thus, the value of iPSCs comes from their ability to be differentiated into cell types that may be otherwise inaccessible for study. iPSC-derived sensory neurons (iPSC-dSNs), for example, can be utilized in high-throughput assays to study diseases affecting the peripheral nervous system, such as peripheral neuropathy, and for the discovery of new drug targets. Reliable differentiation of sensory neurons from patient-specific somatic tissue has the potential to serve as a valuable resource for studying disparities in treatment response and disease progression based on germline genetic factors related to sex, age, or ancestry.
Current reprogramming methods that allow for the derivation of human iPSCs from various somatic tissue types, such as keratinocytes, fibroblasts, and peripheral blood, have greatly advanced neuroscience7,8,9,10,11,12,13,14,15,16,17. However, the implementation of iPSCs as a model is not without limitation. Several studies have demonstrated significant variability in the behavior of reprogrammed iPSC lines18,19,20,21,22,23,24, and specifically in the differentiation of sensory neurons from iPSCs25,26,27,28,29,30. Among various factors that may contribute to this variability, the impact of iPSC passage number is not clearly understood.
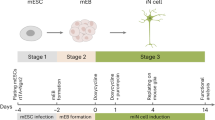
In this study, we examined the effect of iPSC passage number on sensory neuronal differentiation using three lines reprogrammed from the peripheral blood of three donors. We assessed the morphology and pluripotency of each iPSC line at each of three (low, intermediate, and high) passage numbers before differentiation. We then assessed morphology, gene expression, and electrophysiological properties in the resultant iPSC-dSNs to gain a better understanding of the effect of passage number on the derivation of mature, functional sensory neurons.
Results
Passage number does not affect morphology or pluripotency of iPSCs
The morphology and pluripotency of three different iPSC lines (06401-2sb, CMC226, and rsAA-12), each at low (LP), intermediate (IP), and high passage (HP) numbers, were compared. Phase-contrast images demonstrated the expected morphology of flat, well-defined cell colonies in iPSCs of all passage numbers (Fig. 1A). To assess the effect of passage number on the pluripotency of iPSCs, we compared the expression levels of pluripotency markers Sox2, Oct3/4, and Nanog in iPSCs by flow cytometry to determine the proportion of pluripotent cells. Our results indicated that there was no significant difference in the proportion of pluripotent cells across passage number, with an average of 67.5%, 56.9%, and 62.6% of iPSCs expressing all three factors at low, intermediate, and high passage numbers, respectively (Fig. 1B). We further assessed the expression of SOX2, OCT3/4, and NANOG by RNA-seq and the results were consistent; expression of these markers was not significantly different across passage numbers (Fig. 1C). These data indicate that passage number did not affect iPSC morphology or pluripotency.
Morphology and pluripotency of the three iPSC lines (06401-2sb, CMC226, rsAA-12) at different passage numbers (low, intermediate, and high). (A) Phase-contrast images of three iPSC cell lines at different passage numbers. Scale bar = 1000 microns. (B) The average proportion of pluripotent cells in iPSCs at different passage numbers based on measurements of the expression of Sox2, Oct3/4, and Nanog using flow cytometry. (C) The expression level of SOX2, OCT3/4, and NANOG in iPSCs at different passage numbers based on RNA-seq. LP = low-passage, IP = intermediate-passage, HP = high-passage.
Passage number has no effect on morphology and expression of peripherin and βIII-tubulin in iPSC-dSNs
iPSC-dSNs derived from three different iPSC lines at different passage numbers were co-stained for neuronal markers peripherin and βIII-tubulin at day 33 post-induction. Immunofluorescent images of these neurons presented visually similar staining patterns (Fig. 2A). There were also no marked differences in the expression levels of PRPH (peripherin) or TUBB3 (βIII-tubulin) across passage numbers based on RNA-seq, except for a slightly higher expression of TUBB3 in the HP lines than in the LP lines (9.03 vs. 7.98; FDR = 0.005) (Fig. 2B,C). Phase-contrast images revealed similar morphology of iPSC-dSNs across starting iPSC passage numbers (Fig. 2D). Taken together, starting iPSC passage number did not significantly affect the expression of neuronal markers peripherin and βIII-tubulin or neuronal morphology in iPSC-dSNs.
The morphology and expression of neuronal markers peripherin and βIII-tubulin in iPSC-dSNs differentiated from iPSCs at each of low, intermediate, and high passage numbers. (A) Representative immunofluorescent images of neurons stained for neuronal markers peripherin and βIII-tubulin. Scale bar = 200 microns. (B) Average expression levels of TUBB3 in iPSC-dSNs derived from iPSCs with different passage numbers by RNA-seq. (C) Average expression levels of PRPH in iPSC-dSNs derived from iPSCs with different passage numbers by RNA-seq. (D) Representative phase-contrast images of iPSC-dSNs derived from iPSCs with different passage numbers. Scale bar = 200 microns. LP = low-passage, IP = intermediate-passage, HP = high-passage; *FDR = 0.005.
LP iPSCs produce more mature, sensory-like neurons
To assess the maturity and sensory properties of the differentiated neurons, we examined the expression levels of the immature neuron marker gene PAX6 and several mature sensory neuron markers (Fig. 3). Among the three passage number groups from three cell lines, LP iPSC-dSNs had a significantly lower expression of PAX6 compared to both IP (FDR = 0.006) and HP (FDR = 1.1 × 10–4) iPSC-dSNs at day 33 post-induction. These data indicate that the LP iPSC-dSNs achieved greater maturity at this timepoint. More importantly, LP iPSC-dSNs had the highest expression levels of several key mature sensory neuron marker genes at day 33 post-induction, including TRPM8, POU4F3, CALCA, HCN1, RUNX1, NEFH, PIEZO2, SCN9A, and RUNX1, among the three passage groups; the differences in the expression of these marker genes between LP and HP iPSC-dSNs were statistically significant, with FDRs ranging from 0.04 to 1.7 × 10–5, further suggesting greater maturity of the LP iPSC-dSNs. These results indicate that the LP iPSC-dSNs better recapitulated the desired sensory neuron phenotype.
LP iPSCs produce larger, more electrophysiologically mature neurons
Electrophysiological characterization of the iPSC-dSNs derived from different passage numbers demonstrated that LP iPSC-dSNs had a significantly greater average cell size (14.38 ± 1.60 µm) compared to IP (11.75 ± 1.38 µm) and HP (12.67 ± 1.16 µm) iPSC-dSNs (Fig. 4A). The membrane capacitance of LP iPSC-dSNs was 1.6- and 1.5-fold higher than that of IP and HP iPSC-dSNs, respectively (Fig. 4B). LP iPSC-dSNs also had significantly higher sodium current amplitudes and density at some or all voltages from − 40 to 55 mV than IP or HP iPSC-dSNs (Fig. 4C,D). These results suggest that LP iPSC-dSNs demonstrated superior electrophysiological maturity. IP iPSC-dSNs also had significantly higher sodium current amplitudes and density than HP iPSC-dSNs at some or all voltages from − 35 to 0 mV, suggesting that IP iPSC-dSNs, though inferior to LP iPSC-dSNs, also displayed better electrophysiological maturity compared to HP iPSC-dSNs.
Electrophysiological assessment of iPSC-dSNs differentiated from iPSCs at different passage numbers. (A) Average cell size, (B) average membrane capacitance, and (C) average sodium current amplitude and (D) density from whole-cell patch-clamp recordings. Experiments were performed using 16, 14, and 18 iPSC-dSNs of low-, intermediate-, and high-passage, respectively. LP = low-passage, IP = intermediate-passage, HP = high-passage; *p-value < 0.05.
Discussion
Several studies have demonstrated significant variability in neuronal differentiation from iPSCs25,26,27,28,29,30. Factors related to iPSC culture, such as passage number, have been shown to be significant contributors to this variability2,28,31,32,33,34,35,36,37. Some have suggested that differences in differentiation efficiency may be due to an “epigenetic memory” of the tissue of origin19,20,34,38,39,40,41,42,43,44,45. These genetic remnants are thought to be more prominent at lower passages and might be reduced by extended passaging of an iPSC line33,42,46,47,48,49, however, others demonstrated that increased passaging could introduce greater genomic instability and structural variations31,32,50,51,52,53. Tang et al.54 demonstrated that the differentiation of low-passage human neural progenitor cells resulted in a higher percentage of neurons, while higher-passage NPCs gave rise to more glial cells. This would suggest that the passage number of human pluripotent cells on differentiation may be more complex, and the optimization of passage number is necessary depending on the specific endpoint phenotype desired. Our study assessed and compared the quality of peripheral sensory neurons differentiated from iPSCs at different passage numbers to optimize the generation of neurons that better recapitulate the desired peripheral sensory phenotype.
We assessed the relative maturity of iPSC-dSNs based on the expression of several key neuronal markers. PAX6 encodes a key transcription factor in the development of neural tissues and is a marker for neural progenitor cells. During in vivo neuronal differentiation, PAX6 is known to be upregulated shortly after induction and then downregulated as the neurons mature. In our study, LP iPSC-dSNs expressed significantly lower levels of PAX6 at day 33 post-induction as compared to IP and HP iPSC-dSNs, suggesting that the LP cells may have achieved greater maturity by this timepoint. More importantly, LP iPSC-DSNs exhibited higher expression levels of several marker genes for mature sensory neurons, including genes encoding transcription factors (RUNX1 and POU4F3), ion channels (PIEZO2, TRMP8, HCN1, and SCN9A,) cytoskeletal components (NEFH), and the calcitonin gene-related peptide (CGRP)-producing gene, CALCA. Overall, these results strongly indicate that LP iPSCs are more efficient in generating mature sensory neurons. Although iPSC-derived neurons are known for exhibiting a more immature neuron-like phenotype55,56,57,58, our results suggest that this limitation may be at least partially overcome by differentiating iPSCs at lower passage numbers (< 10) and avoiding differentiation at high passage numbers (> 30).
Importantly, however, our study did not indicate that parent iPSC morphology, pluripotency, iPSC-dSN morphology, or expression of the common neuronal markers βIII-tubulin and peripherin, were associated with passage number. We also did not observe any marked differences in morphology or the expression of neuronal markers βIII-tubulin and peripherin in iPSC-dSNs from different passage number groups, indicating that iPSC passage number did not affect the expression of these neuronal markers in the differentiated sensory neurons. Thus, it is not clear whether βIII-tubulin and peripherin are simply not effective determinants of neuronal quality or whether there is a more complicated interplay. While our study focused on the expression of βIII-tubulin and peripherin neuronal markers, another study conducted by Tagliafierro et al.59 evaluated the expression of Lamin proteins in iPSC-derived dopaminergic and cholinergic neurons in order to recapitulate aging processes in the neuronal nuclear architecture.
Generating neurons that are electrophysiologically mature is important for disease-modeling, as many diseases alter electrophysiological properties. Using whole-cell patch-clamp recordings, this study demonstrated that differentiating iPSCs at low passages resulted in sensory neurons with a larger soma size and corresponding greater membrane capacitance. While the biological significance of cell size and capacitance differences are highly debated and difficult to ascertain, they may be important depending on the intended downstream assays using iPSC-dSNs. LP iPSC-dSNs also exhibited greater sodium current amplitude and density, suggesting a lower action potential threshold and greater action potential amplitude, and therefore greater electrophysiological maturity. This supports the superior maturity of the LP cells, along with our expression findings. Similarly, IP iPSC-dSNs exhibited larger sodium currents and presumably greater electrophysiological maturity compared to HP iPSC-dSNs.
Previously, little work has been done to examine the specific effect of iPSC passage number on the differentiation of sensory neurons. In this study, we have demonstrated a simple, effective method for differentiating sensory neurons from iPSCs reprogrammed from human peripheral blood mononuclear cells and have assessed the impact of iPSC passage number on the heterogeneity of these neurons. Here, we demonstrated that, in an iPSC-derived peripheral sensory neuron model, inducing neuronal differentiation at lower passages (< 10) will more consistently produce neurons that achieve greater maturity and electrophysiological properties by day 33. Higher passage iPSCs likely become vulnerable to greater genomic instability, resulting in less efficient differentiation. These findings may be utilized to improve the reliability of disease-modeling and drug discovery related to disorders of the peripheral nervous system. It is important to note that significant variability exists in iPSC-dSN culture with regards to the specific reprogramming and differentiation methods utilized. Therefore, though it may be appropriate to apply the findings of this study more broadly, future studies, such as analyses on the percent of cells successfully differentiated to neurons, differential transcriptomics, or protein-level corroboration of a greater number of genes, are warranted to provide more insight into the underlying mechanisms that contribute to the variability in differentiation of sensory neurons seen across iPSC passage numbers.
Methods
Generation of iPSCs
All donor samples were obtained with informed, written consent under the approval of the Indiana University Institutional Review Boards (IRB) 1106005767 and all experiments were performed in accordance with the relevant guidelines and regulations. Peripheral blood was collected from three individual donors separately and mononuclear cells (PBMCs) were isolated following either the Erythroid Progenitor Reprogramming Kit (STEMCELL Technologies Inc.) protocol (cell lines 06401-2sb and rsAA-12) or using the BD Vacutainer® CPT™ Tube with Sodium Citrate (BD Biosciences) (cell line CMC226) (Fig. 5).
PBMCs isolated with the Erythroid Progenitor Reprogramming Kit continued to be reprogrammed using this protocol. Briefly, erythroid progenitor cells were expanded for seven days in StemSpan™ SFEM II supplemented with 1% StemSpan™ Erythroid Expansion Supplement (STEMCELL Technologies Inc.) before being transfected with Epi5™ episomal vectors (Invitrogen™) containing five reprogramming factors (Oct4, Sox2, Lin28, Klf4, and L-Myc) via nucleofection. Cells were then transitioned into ReproTeSR™ medium (STEMCELL Technologies Inc.) over seven days. After completing the transition, the cells were maintained in ReproTeSR™ medium (STEMCELL Technologies Inc.) and monitored for up to 18 more days until the formation of iPSC colonies.
PBMCs isolated using the BD Vacutainer® CPT™ tube were reprogrammed using the CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Invitrogen™). Briefly, PBMCs were expanded in StemPro™-34 medium (STEMCELL Technologies Inc.) supplemented with 2 mM L-Glutamine and cytokines. After four days of expansion, the PBMCs were transduced with Sendai virus vectors containing the four Yamanaka factors, Oct4, Sox2, Klf4, and c-Myc. Viruses were removed the following day and the transduced cells were maintained on Matrigel-coated plates for an additional seven days, after which they were transitioned into mTeSR™ Plus (STEMCELL Technologies, Inc.) and maintained for up to 20 more days while awaiting the formation of iPSC colonies.
iPSC colonies were then allowed to grow until reaching an appropriate size for selection and transfer. iPSC colonies were then maintained in mTeSR™ Plus medium and tested for pluripotency as described below. Normal karyotypes (46,XX) were confirmed for each line using G-banded cytogenetic analysis.
Maintenance of iPSCs
iPSCs were maintained in mTeSR™ Plus medium on Matrigel-coated plates by a single researcher in order to reduce experimental variability. Medium was replaced daily, and cultures were frequently observed for spontaneous differentiation. Any observed differentiation was removed manually. Each line was passaged one to two times per week using Dispase (1 U/mL in DMEM/F-12; STEMCELL Technologies Inc.) until each target passage number (below) was reached (nine samples total; three cell lines with three target passage numbers each). Low-passage cells were analyzed after 5–10 passages, intermediate-passage cells after 20–26 passages, and high-passage cells after 30–38 passages. The MycoAlert™ Mycoplasma Detection Kit (Lonza) was utilized to confirm the absence of contamination.
Measurement of pluripotency of iPSC lines
The pluripotency of each line was tested via flow cytometry at each of the target passage numbers using the Human Pluripotent Stem Cell Transcription Factor Analysis Kit (BD Biosciences). The expression of three core pluripotency transcription factors (Oct3/4, Sox2, and Nanog) was examined graphically. Unstained and isotype controls were included with every sample.
Neuronal differentiation
iPSCs from the three different cell lines were induced into sensory neuronal differentiation on Matrigel-coated 6-well plates at ~ 80% confluence at each of three different passage numbers (low, intermediate, and high), as described above. Differentiation into sensory neurons (iPSC-dSNs) was achieved via a previously published protocol60 and source information for all small molecules, growth factors, and antibodies used can be found in Table 1. Briefly, differentiation was induced using DMEM/F12 supplemented with KnockOut™ Serum Replacement (10%) (Gibco), LDN-193189 (0.3 µM), A83-01 (2 μM), CHIR 99021 (6 μM), RO4919097 (2 μM), SU 5402 (3 μM), and retinoic acid (0.3 μM). Cells were maintained in this induction medium for eight days, with complete medium replacement occurring every other day. On day nine post-induction, cells were dissociated with Accutase™ (STEMCELL Technologies Inc.). Cells were then strained through a 37 µm mesh and single-cell seeded onto new Matrigel-coated 6-well plates (6 × 10^5 cells/well) in a maintenance medium consisting of Neurobasal™ Plus medium supplemented with B-27 Plus (1%) (Gibco), neurotrophin-3 (10 ng/mL), brain-derived neurotrophic factor (20 ng/mL), nerve growth factor (20 ng/mL), and glial cell line-derived growth factor (20 ng/mL). This maintenance medium was replaced every other day and cells were allowed to mature for up to an additional 24 days.
Phase-contrast imaging
Phase-contrast images of the parent iPSCs and differentiated iPSC-dSNs were captured using the Lionheart FX Automated Microscope (BioTek Instruments, Inc.). Focus and exposure settings were adjusted for optimal visualization. Representative images of the morphology of the three cell lines were captured at either 4X (iPSCs) or 10X (iPSC-dSNs) magnification at low, intermediate, and high passage numbers for visual comparison to assess for any differences in morphology across passage numbers.
Immunofluorescence imaging
On day 28 post-induction, iPSC-dSNs were dissociated using Accutase™ (STEMCELL Technologies, Inc.) and single-cell seeded onto Matrigel-coated 12-well plates at 1.5 × 105 cells/well. The cells were then fixed with paraformaldehyde (4%) on day 33 post-induction and were subsequently stained for immunofluorescence imaging with antibodies against peripherin (1:1000; Abcam) and βIII-tubulin (1:1000; Abcam). Nuclei were stained with NucBlue™ Fixed Cell ReadyProbes™ Reagent (1:1000; Invitrogen™). All staining was carried out by the same researcher in order to reduce variability. Images were captured at 10X magnification using the Lionheart FX Automated Microscope. Representative images for each of the cell lines at different passage numbers were compared visually to assess for any differences in the distribution of the stains or co-staining patterns, as well as morphology.
Electrophysiology
On day 28 post-induction, iPSC-dSNs derived from iPSC line 06401-2sb at each of three passage numbers were single-cell seeded onto 5 × 5 mm Matrigel-coated plastic coverslips at a density of 1.0–1.5 × 105 cells/coverslip. Cell density was adjusted within this range as necessary to achieve proper spacing for patch-clamping. On day 33 post-induction, whole-cell patch-clamp recordings were conducted in voltage-clamp mode at room temperature as previously reported61,62. An Axopatch 200B patch-clamp amplifier (Molecular Devices) was utilized and data were acquired using the pClamp (v8.0) software (Molecular Devices). Borosilicate glass capillaries were used to construct fire-polished electrodes (1.5–2.5 MΩ). The standard electrode solution contained CsF (140 mM), NaCl (10 mM), EGTA (1.1 mM), and HEPES (10 mM, pH 7.3). The standard extracellular bathing solution consisted of NaCl (130 mM), TEA chloride (30 mM), MgCl2 (1 mM), KCl (3 mM), CaCl2 (1 mM), CdCl2 (0.05 mM), HEPES (10 mM), and D-glucose (10 mM; pH 7.3).
RNA-sequencing
Total RNA was extracted from cells on days 0 (iPSCs) and 33 (iPSC-dSNs) post-induction using the RNeasy Mini Kit (QIAGEN). Three cell lines were used at three different passage numbers (low, middle, and high), for a total of 9 iPSC samples and 9 iPSC-dSN samples. Quality and quantity were evaluated with the 2100 Bioanalyzer (Agilent Technologies); the average RIN number across all samples was 9.71. 100 ng of RNA was used for cDNA library preparation, including mRNA purification/enrichment, RNA fragmentation, cDNA synthesis, ligation of index adaptors, and amplification, following the KAPA mRNA Hyper Prep Ki Technical Data Sheet, KR1352–v4.17 (Roche Corporate). The resulting indexed libraries were quantified and assessed for quality using a Qubit™ Fluorometer (Invitrogen™) and the 2100 Bioanalyzer. Multiple libraries were pooled in equal molarity and then were denatured and neutralized. The libraries were loaded onto the NovaSeq 6000 (Illumina, Inc.) sequencer for 100 base-paired sequencing at a concentration of 300 pM. Approximately 30 million reads were generated per library. The quality of the sequencing data was assessed using FastQC (Babraham Bioinformatics) and all sequenced libraries were mapped to the human genome (hg38) with STAR RNA-seq aligner using the parameter “—outSAMmapqUnique 60.” Bamutils (from ngsutils) was utilized to assess the reads distribution across the genome. Uniquely mapped reads were assigned to hg38 refGene genes using featureCounts (from subread) with the parameters “-s 2 -p -q 10”. MultiQC (v1.9) was utilized to summarize quality control results for sequencing and mapping; genes with a CPM > 0.2 in less than 3 samples were removed from further analyses.
Statistical Analyses
Differential expression analysis of pluripotency genes (NANOG, OCT3/4, and SOX2), and neuron marker genes (TUBB3, PRPH, PAX6, TRPM8, POU4F3, CALCA, HCN1, RUNX1, NEFH, PIEZO2, and SCN9A) were conducted separately using three cell lines in each passage number group at day 33 post-induction with a likelihood ratio test using the edgeR package in R (v3.6.2), with the cell line and passage number as independent variables. The Benjamin-Hochberg procedure was used for multiple comparisons correction of the selected genes and a false discovery rate (FDR) less than 0.05 was considered statistically significant.
Raw electrophysiology data was averaged across all cells in each passage group and analyzed using GraphPad Prism v9.0.0. Differences in average cell size and membrane capacitance were assessed via one-way ANOVA. A post-hoc Tukey test was conducted to determine which specific passage number comparisons were statistically different. Average sodium current amplitude and density were assessed, and means were compared using a two-way mixed-model ANOVA and post-hoc Bonferroni test. Corrected p-values less than 0.05 were considered statistically significant. Patch-clamp data was obtained from 16, 14, and 18 cells of low, intermediate, and high passage, respectively.
Data availability
The corresponding author can provide the datasets utilized in this study on reasonable request. The raw gene expression data is available on Gene Expression Omnibus, GSE193571 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193571).
References
Davila, J. C. et al. Use and application of stem cells in toxicology. Toxicol. Sci. 79, 214–223 (2004).
Blelloch, R., Vanere, M., Yen, J. & Ramalho-Santos, M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell 1, 245–247 (2007).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Park, I.-H. et al. Disease-specific induced pluripotent stem cells. Cell 134, 877–886 (2008).
Anson, B. D., Kolaja, K. & Kamp, T. J. Opportunities for human iPSC cells in predictive toxicology. Clin. Pharmacol. Ther. 89, 754–758 (2011).
Dimos, J. T. et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321, 1218–1221 (2008).
Lee, G. et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406 (2009).
Marchetto, M. C. N. et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539 (2010).
Dage, J. L. et al. Pharmacological characterization of ligand- and voltage-gated ion channels expressed in human iPSC-derived forebrain neurons. Psychopharmacology 231, 1105–1124 (2014).
Das, D. K. et al. Genetic and morphological features of human iPSC-derived neurons with chromosome 15q11.2 (BP1-BP2) deletions. Mol. Neuropsychiatry. 1, 116–123 (2015).
Nagy, J. et al. Altered neurite morphology and cholinergic function of induced pluripotent stem cell-derived neurons from a patient with Kleefstra syndrome and autism. Transl. Psychiatry. 7, e1179 (2017).
Wing, C. et al. Application of stem cell derived neuronal cells to evaluate neurotoxic chemotherapy. Stem Cell Res. J. 22, 79–88 (2017).
Bianchi, F. et al. Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling. Stem Cell Res. J. 32, 126–134 (2018).
Yeh, E. et al. Patient-derived iPSCs show premature neural differentiation and neuron-type specific phenotypes relevant to neurodevelopment. Mol. Psychiatry. 23, 1687–1698 (2018).
Dionisi, C., Rai, M., Chazalon, M., Schiffmann, S. N. & Pandolfo, M. Primary proprioceptive neurons from human induced pluripotent stem cells: a cell model for afferent ataxias. Sci. Rep. 10, 7752 (2020).
Robin, G., Evans, J. C., Hauser, D. N., Wren, P. & Zembrzycki, A. Longitudinal characterization of transcriptomic, functional, and morphological features in human iPSC-derived neurons and their application to investigate translational progranulin disease biology. Front. Aging Neurosci. 12, 576678 (2020).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Miura, K. et al. Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 27, 1554 (2009).
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature 467, 09342. https://doi.org/10.1038/nature09342 (2010).
Bock, C. et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439–452 (2011).
Nishizawa, M. et al. Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell 19, 341–354 (2016).
Volpato, V. & Webber, C. Addressing variability in iPSC-derived models of human disease: Guidelines to promote reproducibility. DMM Dis. Models Mech. 13, dmm042317 (2020).
Yamanaka, S. Pluripotent stem cell-based cell therapy – promise and challenges. Cell Stem Cell 27, 523–531 (2020).
Hu, B.-Y. et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 107, 4335–4340 (2010).
Boulting, G. L. et al. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 29, 279–286 (2011).
Koyanagi-Aoi, M. et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc. Natl. Acad. Sci. U. S. A. 110, 20569–20574 (2013).
Volpato, V. et al. Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: a multi-site omics study. Stem Cell Rep. 11, 897–911 (2018).
Burke, E. E. et al. Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat. Commun. 11, 462; https://doi.org/10.1038/s41467-019-14266-z (2020).
Srimasorn, S. et al. Increased neuronal differentiation efficiency in high cell density-derived induced pluripotent stem cells. Stem Cells Int. 2019, 2018784 (2019).
Maitra, A. et al. Genomic alterations in cultured human embryonic stem cells. Nat. Genet. 37, 1099–1103 (2005).
Baker, D. E. C. et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 25, 207–215 (2007).
Chin, M. H., Pellegrini, M., Plath, K. & Lowry, W. E. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell 7, 263–269 (2010).
Mayshar, Y. et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7, 521–531 (2010).
Newman, A. M. & Cooper, J. B. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 7, 258–262 (2010).
Berry, B. J. et al. Morphological and functional characterization of human induced pluripotent stem cell-derived neurons (iCell neurons) in defined culture systems. Biotechnol. Prog. 31, 1613–1622 (2015).
Garitaonandia, I. et al. Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PLoS ONE 10, e0118307 (2015).
Maherali, N. et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70 (2007).
Daley, G. Q. et al. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell 4, 200–201 (2009).
Marchetto, M. C. N. et al. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE 4, e7076 (2009).
Ghosh, Z. et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE 5, e8975 (2010).
Polo, J. M. et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855 (2010).
Bar-Nur, O., Russ, H. A., Efrat, S. & Benvenisty, N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 9, 17–23 (2011).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Ohi, Y. et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13, 541–549 (2011).
Jang, J. et al. Disease-specific induced pluripotent stem cells: a platform for human disease modeling and drug discovery. Exp. Mol. Med. 44, 202–213 (2012).
Hussein, S. M. et al. Copy number variation and selection during reprogramming to pluripotency. Nature 471, 58–62 (2011).
Koehler, K. R. et al. Extended passaging increases the efficiency of neural differentiation from induced pluripotent stem cells. BMC Neurosci. 12, 82 (2011).
Robinton, D. A. & Daley, G. Q. The promise of induced pluripotent stem cells in research and therapy. Nature 481, 295–305 (2011).
Närvä, E. et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat. Biotechnol. 4, 371–377 (2010).
Laurent, L. C. et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8, 106–118 (2011).
Liu, P. et al. Passage number is a major contributor to genomic structural variations in mouse iPSC cells. Stem Cells. 32, 2657–2667 (2014).
Merkle, F. T. et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545, 229–233 (2017).
Tang, X. et al. Astroglial cells regulate the developmental timeline of human neurons differentiated from induced pluripotent stem cells. Stem Cell Res. 11, 743–757 (2013).
Grskovic, M., Javaherian, A., Strulovici, B. & Daley, G. Q. Induced pluripotent stem cells – opportunities for disease modelling and drug discovery. Nat. Rev. 10, 915–929 (2011).
Meneghello, G. et al. Evaluation of established human iPSC-derived neurons to model neurodegenerative diseases. Neurosci. J. 301, 204–212 (2015).
Gunhanlar, N. et al. A simplified protocol for differentiation of electrophysiologically mature neuronal networks from human induced pluripotent stem cells. Mol. Psychiatry. 23, 1336–1344 (2017).
Harbom, L. J. et al. The effect of rho kinase inhibition on morphological and electrophysiological maturity in iPSC-derived neurons. Cell Tissue Res. 375, 641–654 (2019).
Tagliafierro, L., Zamora, M. E. & Chiba-Falek, O. Multiplication of the SNCA locus exacerbates neuronal nuclear aging. Hum. Mol. Genet. 28, 407–421 (2019).
Cai, S., Shum, D. K. Y. & Chan, Y.-S. Human induced pluripotent stem cell-derived sensory neurons for fate commitment of bone marrow stromal cell-derived Schwann cells. In Schwann cells (eds Monje, P. & Kim, H.) 149–160 (Humana Press, 2018).
Tan, Z. Y. et al. Tetrodotoxin-resistant sodium channels in sensory neurons generate slow resurgent currents that are enhanced by inflammatory mediators. J. Neurosci. 34, 7190–7197 (2014).
Su, X. et al. Inhibitory effects of columbianadin on nociceptive behaviors in a neuropathic pain model, and on voltage-gated calcium currents in dorsal root ganglion neurons in mice. Front. Pharmacol. 10, 1522 (2020).
Acknowledgements
We thank our donors for donating blood for this study. We also thank Susan G. Komen for the Cure and the Vera Bradley Foundation for Breast Cancer for supporting this research.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conceptualization and design of the study, data interpretation, and manuscript revisions. E.C. performed experiments, data acquisition, data analysis, and wrote the manuscript. F.S. conducted critical review of the manuscript. G.J. performed data analysis and statistics. Z.T. performed electrophysiology experiments and data analysis. B.P.S. acquired funding for the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cantor, E.L., Shen, F., Jiang, G. et al. Passage number affects differentiation of sensory neurons from human induced pluripotent stem cells. Sci Rep 12, 15869 (2022). https://doi.org/10.1038/s41598-022-19018-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19018-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.