Abstract
Theileria equi, an intraerythrocytic protozoan parasite, causes equine piroplasmosis, a disease which negatively impacts the global horse industry. Genetic manipulation is one of the research tools under development as a control method for protozoan parasites, but this technique needs to be established for T. equi. Herein, we report on the first development of a stable transgenic T. equi line expressing enhanced green fluorescent protein/blasticidin S deaminase (eGFP/BSD). To express the exogenous fusion gene in T. equi, regulatory regions of the elongation factor-1 alpha (ef-1α) gene were identified in T. equi. An eGFP/BSD-expression cassette containing the ef-1α gene promoter and terminator regions was constructed and integrated into the T. equi genome. On day 9 post-transfection, blasticidin-resistant T. equi emerged. In the clonal line of T. equi obtained by limiting dilution, integration of the eGFP/BSD-expression cassette was confirmed in the designated B-locus of the ef-1α gene via PCR and Southern blot analyses. Parasitaemia dynamics between the transgenic and parental T. equi lines were comparable in vitro. The eGFP/BSD-expressing transgenic T. equi and the methodology used to generate it offer new opportunities for better understanding of T. equi biology, with the add-on possibility of discovering effective control methods against equine piroplasmosis.
Similar content being viewed by others
Introduction
Equine piroplasmosis is a tick-borne infectious disease of horses caused by Theileria equi or Babesia caballi. Both parasites are intraerythrocytic haemoprotozoans within the order Piroplasmida (phylum Apicomplexa)1. Equine piroplasmosis often causes huge economic losses to the equine industry, particularly by impairing international trade and sport2,3. T. equi, which is more pathogenic than B. caballi, is widely distributed in tropical and subtropical regions, coinciding with the distribution of its tick vectors4. Horses with acute or sub-acute infection stages of T. equi may develop severe clinical signs, including fever, hemoglobinuria, anemia, and icterus2. Should infected horses recover from this clinical disease, they become carriers of T. equi infection for several years and the parasites persist in many of their organs, including the bone marrow, liver, spleen, lungs, heart, and brain5,6. Vaccine unavailability, coupled with the low efficacy and toxic-side effects of currently used drugs, emergence of drug-resistant parasites, and the rapid development of acaricide resistance in ticks make controlling T. equi infection extremely difficult7,8,9. Hence, although development of improved control measures is vital, it requires a comprehensive understanding of the biology of T. equi.
In common with other Theileria species, T. equi sporozoites, when injected into the host by infected ticks, invade leukocytes where they undergo schizogony to form schizonts in the infected cells10,11,12. When schizonts rupture they release merozoites, which after infecting other red blood cells (RBCs) undergo merogony (asexual reproduction) and produce more merozoites2. These merozoites infect uninfected RBCs, where they continue to proliferate during merogony. Detailed information about invasion, schizont formation, and asexual reproduction is, however, scant for T. equi, and this undermines the development of control and preventive strategies against this parasitic disease. The tick vectors that are competent to be parasitised by T. equi parasites acquire them when feeding on infected animals, and these parasites undergo sexual reproduction in the tick midgut13. As with the asexual stages in the equine host, however, the sexual stages of T. equi—which occur in the tick vector—require thorough investigation. Similar to other Theileria species, T. equi persists transstadially in the subsequent tick stages, which then transmit the infection to other non-infected equines2,12,14. However, transovarial transmission in ticks, which is a unique feature of Babesia, cannot be entirely ruled out for T. equi14. Therefore, research to clarify the developmental stages of T. equi in horses and ticks will be essential for achieving effective control of this economically significant parasite species. The lack of advanced research tools is certainly one of the major constraints in T. equi research.
Genetic manipulation helps researchers to perform experiments wherein the gain or loss of specific gene expression and function is achieved in a pathogen. This can provide researchers with fresh insights into a parasite’s life cycle, pathogenesis and host–parasite interactions, for example. Recently, enhanced green fluorescent protein (eGFP) has been widely used as a reporter for the transfection of apicomplexan parasites, such as T. parva, T. annulata, B. bovis, B. bigemina, B. ovata, B. gibsoni, Plasmodium falciparum, Toxoplasma gondii, and Cryptosporidium parvum15,16,17,18,19,20,21,22,23,24,25. More recently, a fusion gene encoding eGFP/blasticidin S deaminase (eGFP/BSD) was successfully introduced as a reporter and drug-selectable marker for developing stable transgenic B. bovis and B. bigemina lines17,18,24,25. Genetic manipulations using these parasites allowed researchers to clarify the mechanisms underlying cellular invasion, identify vaccine and drug candidates, and conduct cutting-edge research to develop effective preventive and control measures24,25,26.
Despite these advances in apicomplexan research, tools for genetic manipulation of T. equi have not been developed. Hence, we aimed to establish a method for stable transformation of T. equi by integrating the eGFP/BSD gene into this parasite’s genome.
Results
Inhibitory effect of blasticidin on a parental strain of T. equi in vitro
To determine the appropriate blasticidin concentration to use as the selection marker for the eGFP/BSD transgenic parasites, the inhibitory effect of blasticidin was tested on the in vitro growth of the T. equi USDA strain. Blasticidin was found to significantly inhibit the growth of the parental line in a dose-dependent manner, and its half maximal inhibitory concentration (IC50) was determined to be 7.0 ± 0.91 µM (Supplementary Fig. S1).
Construction of a transfer plasmid carrying the eGFP/BSD-expression cassette for use in T. equi
The ef-1α gene’s promoter was used to express the eGFP/BSD gene in T. equi. To identify the promoter region in the T. equi USDA strain’s genome, the mRNA sequence of the ef-1α gene was determined using the rapid amplification of cDNA ends (RACE) technique. Briefly, two amplicons of approximately ~ 0.7- and ~ 1.1-kilo base pairs (kbp) long were obtained from the 5′- and 3′-RACE PCR reactions, respectively (Fig. 1a). These two amplicons were cloned and sequenced. Based on the resultant mRNA sequences from the USDA strain, we identified two intron-lacking loci belonging to the ef-1α gene (1.347-kbp); these two sequences were also found on chromosome 2 of the T. equi WA strain (GenBank accession no. NW_004668230.1; Gene ID no. BEWA_037990 (695,088–696,737) and BEWA_038000 (697,369–698,964), respectively). The two loci in the T. equi WA strain contain two ef-1α head-to-head-oriented ORFs flanked by ribonucleoside-diphosphate reductase (rdrs; Gene ID: BEWA_037980) and glutamyl-tRNA synthetase (gluRS; Gene ID: BEWA_038010) genes. Therefore, we used a pair of forward and reverse primers targeting the rdrs and gluRS genes, respectively, to amplify a fragment of ~ 5.46-kbp from the genomic DNA (gDNA) of the T. equi USDA strain. The amplicon was cloned and sequenced (Fig. 1b). Our analysis confirmed that the amplified DNA fragment contained ef-1α-A gene, 1.164-kbp intergenic region (IG) containing 0.246-kbp inverted repeats at its both ends, and ef-1α-B gene (Fig. 1c). Based on the sequence, the promoter (0.87-kbp of the 1.164-kbp IG located closer to the ef-1α-B gene), terminator (0.278-kbp of the region located between rdrs and ef-1α-A genes) and two homologues flanking segments (0.628- and 0.719-kbp of the regions located at the 5′ and 3′ ends of the ef-1α-B gene, respectively) were PCR-amplified and then inserted into a Bluescript plasmid (Fig. 1d). In addition, to express the reporter gene and drug selectable marker in T. equi, a 1.104-kbp eGFP/BSD fusion gene amplified from the pTracer-CMV/Bsd Mammalian Expression Vector was inserted between the promoter and terminator segments in the constructed Bluescript plasmid described above. The nucleotide sequence of the eGFP/BSD-expression cassette for T. equi is deposited in GenBank (Accession No. MW021139). The eGFP/BSD-expression cassette was then targeted to the ef-1α-B gene locus in the genome of T. equi (Fig. 1e).
Construction of the enhanced green fluorescent protein/blasticidin S deaminase (eGFP/BSD)-expression cassette targeting the elongation factor-1α-B gene locus in T. equi genome. (a) Visualisation of the amplicons obtained from 5′-RACE PCR (5′) and 3′-RACE PCR (3′). M, 100 base-pair DNA ladder marker. (b) Structure of head-to-head orientation of two copies of elongation factor-1α (ef-1α) genes in the T. equi (USDA strain) genome. The ef-1α gene loci are flanked by ribonucleoside-diphosphate reductase (rdrs) and glutamyl-tRNA synthetase (gluRS) genes. Regions containing the terminator (ter), a part of the intergenic region (IG), homologous flanking fragment A (flank A), and homologous flanking fragment B (flank B) that were selected for plasmid construction are highlighted in grey, orange, blue, and yellow, respectively. (c) Magnified structure of the IG containing inverted region-A (IR-A), non-inverted region, and inverted region-B (IR-B). (d) The eGFP/BSD-expression cassette. The expression cassette included green fluorescent protein (gfp) and blasticidin S deaminase (bsd) genes. (e) Genomic integration of the eGFP/BSD-expression cassette into the ef-1α-B gene locus of T. equi.
Transfection and isolation of a transgenic eGFP/BSD-expressing T. equi line
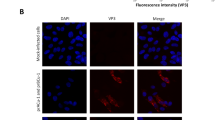
T. equi was transfected with the linearised eGFP/BSD-expression cassette and then treated with 70-μM blasticidin (10 × IC50). Blasticidin-resistant parasites emerged 9 days after transfection (Fig. 2a). On day 12 post-transfection, a green fluorescent signal emission was observed in the parasite’s body (Supplementary Fig. S2). Subsequently, a clonal line of transgenic T. equi was isolated from the initial transfected parasites, using a limiting dilution method conducted under 70-μM blasticidin drug pressure. Even after the blasticidin pressure was discontinued, this green fluorescent signal could be observed in the transgenic clone for at least one year (Fig. 2b). When the transgenic and parental lines were treated with 70-μM blasticidin, the initial parasitaemia (~ 1%) in the transgenic line reached up to 11% within 96 h of treatment, whereas the parasitaemia decreased to below 0.1% in the parental line (Fig. 3a). The parasitaemia curve of the transgenic line was comparable to that of the parental T. equi line when both lines were cultured without blasticidin (Fig. 3b).
Transfection and isolation of the enhanced green fluorescent protein/blasticidin S deaminase (eGFP/BSD)-expressing transgenic T. equi parasite. (a) Selecting the T. equi transgenic line using blasticidin pressure. Blasticidin treatment was initiated on day 1 post-transfection. Blasticidin-resistant T. equi emerged on day 9 post-transfection. (b) Visualisation of T. equi at one year after the isolation of clonal line. A green fluorescent signal was emitted in both the merozoite (ring, paired, and Maltese cross forms) and trophozoite (ring form) stages of the transfected parasites.
Parasitaemia curves of the transgenic T. equi line expressing enhanced green fluorescent protein/blasticidin S deaminase (eGFP/BSD) and the parental line. (a) Transgenic T. equi was resistant to blasticidin but the parental line was not when cultured in medium containing 70-µM blasticidin. (b) The parasitaemia dynamics were comparable between the transgenic and parent lines when cultured in normal medium.
Confirmation of genomic integration of the eGFP/BSD-expression cassette in transgenic T. equi
To confirm that genomic integration of the eGFP/BSD-expression cassette (2.264-kbp: ef-1α gene promoter, coding region of the eGFP/BSD gene and ef-1α gene terminator) had occurred in the transgenic line, PCR and Southern blot analyses were conducted to detect specific amplicons or length polymorphisms in the genome (Supplementary Table S1). When a set of forward and reverse primers targeting the homologous flanking A and B fragments was used for PCR, 0.446- and 2.722-kbp amplicons were detected as arising from the gDNA of the transgenic line, indicating that the eGFP/BSD-expression cassette had integrated only into one of the two loci of the ef-1α genes (Fig. 4a). By contrast, the same PCR primers only amplified the 0.446-kbp fragment from the gDNA of the parental line. Furthermore, when primers targeting rdrs and the homologous flanking fragment A of the ef-1α genes were used in a PCR, a 2.010-kbp fragment was amplified from the gDNA of both transgenic and parental lines (Fig. 4b, left), indicating that eGFP/BSD-expression cassette has not been integrated into the ef-1α-A gene locus which is located close to the rdrs gene. Conversely, when a primer pair targeting the eGFP/BSD and gluRS genes was used in PCR, a 2.759-kbp fragment was amplified from the gDNA of the transgenic line but not from the parental line (Fig. 4b, right). These results confirm that in the transgenic line, the eGFP/BSD-expression cassette had only integrated into the ef-1α-B gene locus located close to the gluRS gene. Moreover, our PCR assays conducted with gDNA from the transgenic line using forward primers that target flanking region A, the promoter, the eGFP/BSD coding region, the terminator, and flanking region B, together with the reverse primer targeting the gluRS gene, resulted in 3.952-, 3.642-, 2.759-, 1.652-, and 1.368-kbp amplicons, respectively (Fig. 4c). In contrast, when the gDNA of the parental line was used, PCRs targeting flanking region A and flanking region B of the ef-1α and gluRS genes amplified 1.676- and 1.368-kbp fragments, respectively, while no amplicon was detected in the remaining three PCRs. Overall, these PCR results further confirm the correct orientation of the eGFP/BSD-expression cassette and its integration into the ef-1α-B gene locus of the transgenic line.
Graphical illustration of genomic integration of the enhanced green fluorescent protein/blasticidin S deaminase (eGFP/BSD)-expression cassette in transgenic T. equi. Genomic DNA extracted from the transgenic T. equi line (TL) and parental line (PL) was subjected to PCR assays. (a) A PCR assay (P1) confirmed the integration of the eGFP/BSD-expression cassette into one of the two elongation factor-1α (ef-1α) gene loci. (b) Two PCR assays (P2 and P3) confirmed that integration of the eGFP/BSD-expression cassette had occurred in the ef-1α-B gene locus. (c) Five PCR assays (P4–P8) confirmed the integration and correct orientation of the eGFP/BSD-expression cassette components in the ef-1α-B gene locus of the transgenic T. equi. A 1000 base-pair DNA ladder marker was used in all panels.
We prepared two types of DNA probes for Southern blot analyses based on the eGFP/BSD gene and promotor region, which detected a 4.976-kbp fragment in the EcoRV restriction enzyme-digested gDNA of the transgenic line (Fig. 5a–c, and Supplementary Fig. S3 and S4). This indicates that the eGFP/BSD-expression cassette had only integrated into a single genomic locus. By contrast, in the Southern blotting experiment that used similarly digested gDNA from the parental T. equi line, a single 2.994-kbp fragment was detected by the promotor-targeting probe, but no band was detected by the probe targeting the eGFP/BSD gene (Fig. 5a,b,d, and Supplementary Fig. S3 and S4). In Western blot performed with a lysate from transgenic T. equi, an anti-GFP rabbit polyclonal antibody recognized a 40-kilodalton protein, corresponding to the size of eGFP/BSD, whereas no protein was detected when lysates from parental line and uninfected equine erythrocytes were analysed (Fig. 5e). Overall, our findings indicate that the eGFP/BSD-expression cassette successfully integrated only into the ef-1α-B gene locus in T. equi, and that the newly generated transgenic line stably expresses eGFP/BSD.
Graphical illustration of Southern blot and Western blot analyses. Genomic DNA extracted from the transgenic T. equi line (TL) and parental line (PL) was subjected to Southern blotting analyses after digestion with EcoRV. (a) A probe targeting the green fluorescent protein gene (GFP-probe) detected a single DNA fragment (4.9-kbp) in the genomic DNA (gDNA) obtained from the transgenic TL, but no such fragment was detectable in the PL. (b) A probe targeting the promoter region of the ef-1α gene (Prom-probe) detected a single 4.97-kbp fragment in the gDNA from the transgenic TL, whereas a 2.99-kbp fragment was detected in the PL. (c) Graphical illustration of ef-1α gene loci in the genome of the transgenic TL and EcoRV recognition sites. (d) Graphical illustration of ef-1α gene loci in the PL genome and EcoRV recognition sites. (e) In Western blot, an anti-GFP rabbit polyclonal antibody recognized a 40-kilodalton protein, corresponding to the size of eGFP/BSD, in lysate of TL but not in the lysates of PL and uninfected erythrocytes (UE).
Discussion
Successful cell transformation necessitates a cell-specific element that can bind to essential transcription factors and function as a strong promoter to drive exogenous genes into the target cells27. A suitable-for-transformation promoter region has not been identified in T. equi. Previous studies have reported that an intergenic region (IG) in the ef-1α gene contains strong promotor activity in piroplasma parasites such as T. parva, B. bovis, B. bigemina, B. ovata and B. gibsoni15,19,28,29,30. Therefore, we opted to use the ef-1α gene’s promoter, which successfully drove expression of the exogenous eGFP/BSD gene in T. equi. The ef-1α is a multi-copy gene in most eukaryotic cells; it encodes a regulatory protein, named as EF-1α, which is involved in translocating aminoacyl-tRNA to the ribosome31. Two ef-1α gene copies exist in the genomes of Plasmodium species and several Babesia species, including B. bovis, B. bigemina, B. ovata and B. gibsoni19,20,28,29,32. Previous studies have shown that the IG region functions as a bidirectional promoter and drives the expressions of the two ef-1α genes in B. bovis, B. bigemina and B. ovata19,28,29. In the present study, a similar bidirectional head-to-head-orientation of the two ef-1α gene copies was identified in the T. equi genome. In contrast, only a single copy of the ef-1α gene was reported in T. annulata, T. parva and T. orientalis (Gene IDs: TA06720, TP01_0726, TOT_010000685, respectively; https://piroplasmadb.org/piro/)33,34. Thus, generating transgenic parasites by integrating exogenous genes into the ef-1α gene locus is impossible in the aforementioned bovine Theileria parasites.
In B. bigemina, knocking out one of the two ef-1α gene copies resulted in a 3-times higher parasitaemia level in the transgenic line in comparison with that of the parental line, whereas the parasitaemia levels of transgenic B. bovis and its parental line were indistinguishable18. Similar to that found in B. bovis, knocking out the ef-1α-B gene in T. equi did not alter the parasitaemia level, when compared with that of the parental line. These findings suggest that our newly-developed transgenic T. equi is similar to the parental line in that it can invade, replicate and egress.
Our microscopic observations on the transgenic T. equi revealed that eGFP/BSD was stably expressed in all of its intraerythrocytic stages. A previous study demonstrated that ef-1α gene promoter stably drives expression of exogenous genes in P. falciparum throughout its life cycle21. Therefore, the transgenic T. equi may also express eGFP/BSD in all parasite stages in the equine host and tick vectors. However, only the experimental infections with transgenic T. equi will confirm our assumption.
The transgenic T. equi line developed herein will enable better visualisation of live parasites, and this is useful because the developmental stages of T. equi have not currently been investigated thoroughly enough in host blood cells. The lack of schizonticidal drugs makes complete clearance of T. equi from infected horses very difficult, and research on drug development is severely constrained because the formation and persistence of T. equi schizonts in infected horses awaits proper elucidation. Current knowledge on RBC invasion and asexual reproduction in T. equi is also incomplete. Notably, the invasion process, which is aided by the gliding motility of Babesia merozoites, is largely unknown in T. equi. The transgenic T. equi newly generated in the present study offers a powerful research tool for investigating the developmental stages of the parasite in its host blood cells. Its use provides opportunities for gaining fresh insight into schizont formation, RBC invasion, and asexual reproduction.
Vaccination against the equine piroplasmosis caused by T. equi would be one useful control method for minimizing economic losses in the equine industry1,2. However, to date, no vaccines are available for equine piroplasmosis. Recently, several antigens have been evaluated as vaccine candidates against various haemoprotozoan diseases1,35,36,37. Generally, such candidates are usually screened in vitro for their neutralisation sensitivities using antisera produced against them35. However, this strategy provides indirect evidence and has sometimes proved unsuitable because the candidate antigens can elicit non-specific immunity24. In contrast, gene knockout can provide a definitive answer as to whether the antigens under investigation are indispensable for parasite survival. Therefore, the methodology that we have used to develop transgenic T. equi would greatly facilitate the identification of vaccine candidates against equine piroplasmosis. In addition to the potential application in vaccine development, this technique may also be useful for discovering novel drug targets in T. equi.
In summary, we have developed the first stable eGFP/BSD-expressing transgenic T. equi line. Our transgenic T. equi and the methodology used to generate it have potential to augment current understanding of T. equi biology and further the development of effective control and preventive measures against equine piroplasmosis.
Methods
In vitro cultures of T. equi
T. equi (USDA strain) was maintained in vitro in an atmosphere 5% O2 and 5% CO2 at 37 °C, as previously described38. Briefly, T. equi was grown in purified horse red blood cells (RBCs) using M199 medium supplemented with 40% horse serum and hypoxanthine (Sigma-Aldrich, Tokyo, Japan) at 13.6 μg/ml. An antibiotic–antimycotic solution, containing 60 U/ml penicillin G, 60 μg/ml streptomycin, and 0.15 μg/ml amphotericin B (Sigma-Aldrich, Tokyo, Japan), was also added at a final concentration of 1% (v/v).
Blasticidin susceptibility of T. equi
An in vitro growth inhibition assay was conducted to determine the half-maximal inhibitory concentration (IC50) of blasticidin against T. equi, as previously described39. Briefly, T. equi cultures at 10% hematocrit and 1% initial parasitaemia were treated with blasticidin at 0.8, 1.6, 3.1, 6.3, 12.5, 25, and 50 µM for 4 days. The medium, which contained 1% Milli-Q water that was equal to the volume of Milli-Q water used to dissolve the highest concentration of blasticidin, served as the non-treated control. The parasitaemias, which were determined 96 h after cultivation by counting the number of infected RBCs among > 5,000 RBCs in the Giemsa-stained RBC smears using a light microscope, were converted to growth rates relative to the non-treated control. IC50 values were calculated using GraphPad Prism (GraphPad Software, San Diego, CA, USA). These experiments were conducted in triplicate and repeated thrice.
Sequencing ef-1α mRNA from T. equi
Total RNA was extracted from 200 μl of T. equi-infected RBCs using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA was purified using the QIAamp RNA Blood Mini Kit (Qiagen, Tokyo, Japan) with recombinant DNase I digestion (Takara, Otsu, Japan). This RNA was subjected to cDNA synthesis using the SMARTer RACE cDNA Amplification Kit (Takara, Kyoto, Japan). Forward (5′-GATTACGCCAAGCTTCGTGAGCATGCCTTGTTGGCCTTC -3′) and reverse primers (5′-GATTACGCCAAGCTTGGTCCTCCTTGTAGTCGCACTTG-3′), which were designed according to the mRNA sequences of the ef-1α genes from T. equi (WA strain, XM_004833157.1), B. bovis (AK441624.1), B. bigemina (KT439182.1), and T. parva (XM_761154.1), were used in 5′-RACE PCR and 3′-RACE PCR, respectively. The amplified PCR products were then cloned into a pRACE vector, transformed into Escherichia coli (Stellar Competent Cells) using the In-Fusion HD Cloning Kit (Takara, Kyoto, Japan), and then sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Tokyo, Japan). The sequences of the 5′-RACE and 3′-RACE PCR products were assembled and analysed using Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) software and the EMBOSS needle pairwise sequence alignment tool (https://www.ebi.ac.uk/Tools/psa/emboss_needle/).
Amplification and sequencing of long DNA fragments of elongation factor-1 alpha locus
gDNA was extracted from 200 µl of T. equi-infected RBCs using the QIAamp DNA Blood Mini Kit (Qiagen, Tokyo, Japan) with RNase A digestion (Takara, Otsu, Japan), according to the manufacturer’s instructions. The entire ef-1α gene locus (5.46-kbp) was amplified using a set of forward and reverse PCR primers designed against rdrs (5′-TCAGCCAAAATAGCGCACAAAAACTCCC-3′) and gluRS (5′-CCCAAAGCAACCCAAGCCATCTATG-3′) genes, respectively. KOD Fx Neo DNA polymerase, PCR buffer for KOD FX Neo and dNTPs (Toyobo, Osaka, Japan) were used according to the manufacturer’s instructions. The PCR amplicon was cloned into pCR 4Blunt-TOPO vector, transformed into TOP10 E. coli (Invitrogen, Carlsbad, CA, USA), and then sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems).
Transfer plasmid construction
The homologous A and B flanking fragments and the promoter and terminator regions of the ef-1α gene were amplified from the 5.46-kbp fragment-containing pCR 4Blunt-TOPO vector using the PCR primers listed in Supplementary Table S2. The coding region of eGFP/BSD was also amplified from the pTracer-CMV/Bsd Mammalian Expression Vector (Invitrogen, Carlsbad, CA, USA). Homologous flanking fragments A and B were inserted into a Bluescript plasmid after digestion with SacI and XmaI restriction enzymes (New England Biolabs, Ipswich, MA, USA), respectively, using the In-Fusion HD Cloning Kit (Takara, Kyoto, Japan), followed by insertion of the XbaI-digested terminator region of the ef-1α gene between the A and B fragments. In parallel, the ef-1α gene’s promoter sequence and the eGFP/BSD coding region were inserted into a second Bluescript plasmid after digestion with SacI or XbaI, respectively. The fragment containing the promoter and the eGFP/BSD coding region was then amplified from the second plasmid, digested with NarI, and inserted into the first plasmid between flanking fragment A and the terminator. The constructed plasmid containing the eGFP/BSD-expression cassette was purified using Qiagen Plasmid Maxi Kit (Qiagen, Tokyo, Japan), according to the manufacturer’s instructions.
Parasite transfections
The eGFP/BSD-expression cassette-containing transfer plasmid was digested with ApaLI (New England Biolabs) before its transfection into T. equi. At 10% parasitaemia, the T. equi-infected RBCs were washed twice with phosphate-buffered saline. Linearised plasmids (20 μg) were resuspended in 100 µl of nucleofector human T-cell solution (Lonza, Cologne, Germany) and then mixed with pre-washed, 100-µl-infected RBCs. T. equi was then transfected with the linearised plasmid via a Nucleofector 2b device (Lonza) using the V-024 program. The transfectant was then immediately added to 1 ml of culture medium containing fresh RBCs at 10% hematocrit. At 24 h post-transfection, the culture containing transfected T. equi was treated daily with blasticidin at 70-µM (10 × IC50). Two weeks later, a single clonal line was isolated by limiting dilution. Infected RBCs (1 μl) from the clonal line were used for fluorescence microscopic observations (1,000 × magnification, Keyence, Pasadena, CA, USA). Parasite nuclei were stained with Hoechst 33,342 (Invitrogen, Carlsbad, CA, USA).
Confirming genomic integration and expression of the eGFP/BSD
Genomic integration of eGFP/BSD-expression cassette was confirmed using PCR assays and Southern blot analyses. The PCR assays were conducted using gDNAs extracted from transgenic and parental lines with forward and reverse primers (Supplementary Table S1) targeting homologous flanking fragments A and B, eGFP/BSD and the gluRS gene, and eGFP/BSD and the rdrs gene, respectively. PCR assays using forward primers targeting flanking region A, promoter, eGFP/BSD coding region, terminator and flanking region B, together with the gluRS-targeting reverse primer were also conducted. The presence or absence of PCR amplicons and their sizes were compared between the transgenic and parental lines.
For the Southern blot analyses, gDNAs (2 µg) extracted from the transgenic and parental lines were digested with EcoRV restriction enzyme, electrophoresed on a 0.8% agarose gel, stained with Midori Green Advance DNA-staining (Nippon Genetics, Tokyo, Japan), and then transferred onto Hybond N+ (GE Health-care, Buckinghamshire, UK). Two DNA probes targeting the eGFP/BSD coding or promotor region were prepared using two sets of forward (5′-CCAAAGGAGAAGAACTTTTCACTGG-3′ or 5′-TTTGTGTCCGAGAATGTTTCC-3′, respectively) and reverse (5′-CCAAAGGAGAAGAACTTTTCACTGG-3′ or 5′-TTTGTGTCCGAGAATGTTTCC-3′, respectively) primers. The probes were labelled with thermostable alkaline phosphatase using the Alkphos Direct kit (GE Healthcare), according to the manufacturer’s instructions. Reactions between the labelled probes and DNA were detected using CDP-star detection reagent (GE Healthcare).
We also confirmed the expression of eGFP/BSD in transgenic T. equi in Western blot assay. Briefly, 180 µL of parasite-infected erythrocytes with ~ 5% to 8% parasitaemia were treated with 0.15% saponin, and then lysed in phosphate buffered-saline containing 1% Triton X-100 (Sigma-Aldrich, Tokyo, Japan) and protease inhibitor (Roche Diagnostics, Mannheim, Germany). The lysate was separated in 12.5% acrylamide gel, and then transferred onto a nitrocellulose membrane (Bio-Rad Laboratories, Feldkirchen, Germany). The membrane was blocked using 5% skim milk (Wako, Osaka, Japan), stained with anti-GFP rabbit polyclonal antibody at 1:500 (v/v) dilution (Thermo Fisher Scientific, MA, USA), and then incubated with horseradish peroxidase-conjugated anti-rabbit IgG donkey polyclonal antibody at 1:10,000 (v/v) dilution (GE Healthcare). The reaction was detected with Amersham ECL Western Blotting Detection Reagents (GE Healthcare) using ImageQuant LAS 500 chemiluminescence CCD camera system (GE Healthcare), according to the manufacturer’s instructions.
In vitro growth rate assessments
The transgenic and parental lines (1% parasitaemia) were cultured in vitro using horse RBCs at 10% haematocrit. The cultures were treated with 70-µM blasticidin for 4 days. In addition, the transgenic and the parental lines were grown in normal medium without blasticidin. Parasitaemia was microscopically monitored using Giemsa-stained RBC smears prepared every 24 h until 96 h post-cultivation. This experiment was conducted in triplicate and repeated three times.
Ethics approval
All experiments were approved by the Animal Care and Use Committee and the Biological Safety Committee of Obihiro University of Agriculture and Veterinary Medicine, Hokkaido, Japan (Approval numbers: 18-118) and were conducted according to the Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Data availability
Any requests for materials should be addressed to Naoaki Yokoyama (DVM, PhD); E-mail: yokoyama@obihiro.ac.jp.
References
Kappmeyer, L. S. et al. Comparative genomic analysis and phylogenetic position of Theileria equi. BMC Genomics 13, 603. https://doi.org/10.1186/1471-2164-13-603 (2012).
Wise, L. N., Kappmeyer, L. S., Mealey, R. H. & Knowles, D. P. Review of equine piroplasmosis. J. Vet. Intern. Med. 27, 1334–1346. https://doi.org/10.1111/jvim.12168 (2013).
Knowles, D. Jr. Equine babesiosis (piroplasmosis): a problem in the international movement of horses. Br. Vet. J. 152, 123–126. https://doi.org/10.1016/s0007-1935(96)80066-2 (1996).
Scoles, G. A. & Ueti, M. W. Vector ecology of equine piroplasmosis. Annu. Rev. Entomol. 60, 561–580. https://doi.org/10.1146/annurev-ento-010814-021110 (2015).
Ribeiro, I. B. et al. Detection of Theileria equi in spleen and blood of asymptomatic piroplasm carrier horses. Acta Parasitol. 58, 218–222. https://doi.org/10.2478/s11686-013-0127-9 (2013).
Alhassan, A. et al. Comparative evaluation of the sensitivity of LAMP, PCR and in vitro culture methods for the diagnosis of equine piroplasmosis. Parasitol. Res. 100, 1165–1168. https://doi.org/10.1007/s00436-006-0430-6 (2007).
Adams, L. G. Clinicopathological aspects of imidocarb dipropionate toxicity in horses. Res. Vet. Sci. 31, 54–61. https://doi.org/10.1016/S0034-5288(18)32521-9 (1981).
Hines, S. A. et al. Theileria equi isolates vary in susceptibility to imidocarb dipropionate but demonstrate uniform in vitro susceptibility to a bumped kinase inhibitor. Parasit. Vectors 8, 33. https://doi.org/10.1186/s13071-014-0611-6 (2015).
Kunz, S. E. & Kemp, D. H. Insecticides and acaricides: resistance and environmental impact. Rev. Sci. Tech. 13, 1249–1286. https://doi.org/10.20506/rst.13.4.816 (1994).
Mehlhorn, H. & Schein, E. Redescription of Babesia equi Laveran, 1901 as Theileria equi Mehlhorn, Schein 1998. Parasitol. Res. 84, 467–475. https://doi.org/10.1007/s004360050431 (1998).
Ramsay, J. D. et al. Lymphocytes and macrophages are infected by Theileria equi, but T cells and B cells are not required to establish infection in vivo. PLoS ONE 8, e76996. https://doi.org/10.1371/journal.pone.0076996 (2013).
Shaw, M. K. Cell invasion by Theileria sporozoites. Trends Parasitol. 19, 2–6. https://doi.org/10.1016/s1471-4922(02)00015-6 (2003).
Zapf, F. & Schein, E. The development of Babesia (Theileria) equi (Laveran, 1901) in the gut and the haemolymph of the vector ticks, Hyalomma species. Parasitol. Res. 80, 297–302. https://doi.org/10.1007/BF02351869 (1994).
Jalovecka, M., Hajdusek, O., Sojka, D., Kopacek, P. & Malandrin, L. The complexity of piroplasms life cycles. Front. Cell. Infect. Microbiol. 8, 248. https://doi.org/10.3389/fcimb.2018.00248 (2018).
Goeyse, I. D. et al. Transfection of live, tick derived sporozoites of the protozoan Apicomplexan parasite Theileria parva. Vet. Parasitol. 208, 238–241. https://doi.org/10.1016/j.vetpar.2015.01.013 (2015).
Adamson, R. et al. Transient transfection of Theileria annulata. Mol. Biochem. Parasitol. 114, 53–61. https://doi.org/10.1016/s0166-6851(01)00238-9 (2001).
Suarez, C. E. & McElwain, T. F. Stable expression of a GFP-BSD fusion protein in Babesia bovis merozoites. Int. J. Parasitol. 39, 289–297. https://doi.org/10.1016/j.ijpara.2008.08.006 (2009).
Silva, M. G., Knowles, D. P., Mazuz, M. L., Cook, B. M. & Suarez, C. E. Stable transformation of Babesia bigemina and Babesia bovis using a single transfection plasmid. Sci. Rep. 8, 6096. https://doi.org/10.1038/s41598-018-23010-4 (2018).
Hakimi, H. et al. Establishment of transient and stable transfection systems for Babesia ovata. Parasit. Vectors 9, 171. https://doi.org/10.1186/s13071-016-1439-z (2016).
Liu, M. et al. Establishment of a stable transfection system for genetic manipulation of Babesia gibsoni. Parasites Vectors 11, 260. https://doi.org/10.1186/s13071-018-2853-1 (2018).
Talman, A. M., Blagborough, A. M. & Sinden, R. E. A Plasmodium falciparum strain expressing GFP throughout the parasite’s life-cycle. PLoS ONE 5, e9156. https://doi.org/10.1371/journal.pone.0009156 (2010).
Nishikawa, Y. et al. Construction of Toxoplasma gondii bradyzoite expressing the green fluorescent protein. Parasitol. Int. 57, 219–222. https://doi.org/10.1016/j.parint.2007.10.004 (2008).
Li, W. et al. Transient transfection of Cryptosporidium parvum using green fluorescent protein (GFP) as a marker. Mol. Biochem. Parasitol. 168, 143–148. https://doi.org/10.1016/j.molbiopara.2009.07.003 (2009).
Alzan, H. F. et al. Geno- and phenotypic characteristics of a transfected Babesia bovis 6-Cys-E knockout clonal line. Parasit. Vectors 10, 214. https://doi.org/10.1186/s13071-017-2143-3 (2017).
Hussein, H. E. et al. The Babesia bovis hap2 gene is not required for blood stage replication, but expressed upon in vitro sexual stage induction. PLoS Negl. Trop. Dis. 11, e0005965. https://doi.org/10.1371/journal.pntd.0005965 (2017).
Suarez, C. E., Bishop, R. P., Alzan, H. F., Poole, W. A. & Cooke, B. M. Advances in the application of genetic manipulation methods to apicomplexan parasites. Int. J. Parasitol. 47, 701–710. https://doi.org/10.1016/j.ijpara.2017.08.002 (2017).
Narlikar, L. & Ovcharenko, I. Identifying regulatory elements in eukaryotic genomes. Brief. Funct. Genomics Proteomics 8, 215–230. https://doi.org/10.1093/bfgp/elp014 (2009).
Suarez, C. E., Norimine, J., Lacy, P. & McElwain, T. F. Characterization and gene expression of Babesia bovis elongation factor-1α. Int. J. Parasitol. 36, 965–973. https://doi.org/10.1016/j.ijpara.2006.02.022 (2006).
Silva, M. G., Knowles, D. P. & Suarez, C. E. Identification of interchangeable cross-species function of elongation factor-1 alpha promoters in Babesia bigemina and Babesia bovis. Parasit. Vectors 9, 576. https://doi.org/10.1186/s13071-016-1859-9 (2016).
Liu, M. et al. Transient transfection of intraerythrocytic Babesia gibsoni using elongation factor-1 alpha promoter. Mol. Biochem. Parasitol. 216, 56–59. https://doi.org/10.1016/j.molbiopara.2017.07.003 (2017).
Andersen, G. R. et al. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Mol. Cell 6, 1261–1266. https://doi.org/10.1016/s1097-2765(00)00122-2 (2000).
Vinkenoog, R. et al. Malaria parasites contain two identical copies of an elongation factor 1 alpha gene. Mol. Biochem. Parasitol. 94, 1–12. https://doi.org/10.1016/s0166-6851(98)00035-8 (1998).
Gardner, M. J. et al. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science 309, 134–137. https://doi.org/10.1126/science.1110439 (2005).
Hayashida, K. et al. Comparative genome analysis of three eukaryotic parasites with differing abilities to transform leukocytes reveals key mediators of Theileria-induced leukocyte transformation. MBio 3, e00204-12. https://doi.org/10.1128/mBio.00204-12 (2012).
Silva, M. G. et al. Babesia bovis expresses Bbo-6cys-E, a member of a novel gene family that is homologous to the 6-cys family of Plasmodium. Parasitol. Int. 60, 13–18. https://doi.org/10.1016/j.parint.2010.09.004 (2011).
Liu, Y. L. et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 22, 1051–1068. https://doi.org/10.1101/gad.1656508 (2008).
Barriga, O. O. A review on vaccination against protozoa and arthropods of veterinary importance. Vet. Parasitol. 55, 29–55. https://doi.org/10.1016/0304-4017(94)90054-X (1994).
Averzed, A. et al. Monoclonal antibody against Babesia equi: characterization and potential application of antigen for serodiagnosis. J. Clin. Microbiol. 36, 1835–1839. https://doi.org/10.1128/JCM.36.7.1835-1839.1998 (1998).
Tuvshintulga, B. et al. Clofazimine inhibits the growth of Babesia and Theileria parasites in vitro and in vivo. Antimicrob. Agents Chemother. 60, 2739–2746. https://doi.org/10.1128/AAC.01614-15 (2016).
Acknowledgements
This work was financially supported by the Japan Society for the Promotion of Science (JSPS) (KAKENHI Grant 18H02337, 19KK0174, and 18F18399). B. Tuvshintulga is a post-doctoral fellow (Standard P18399) of the JSPS. Funders played no role in the experimental design, data analysis or manuscript preparation.
Author information
Authors and Affiliations
Contributions
B.T., A.B.N., N.Y. and I.I. conceived the study. A.B.N. and B.T. performed all the experiments. M.L. and X.X. assisted with eGFP/BSD-expression cassette construction; T.I. assisted with Southern blotting analysis; T.S. assisted with Western blotting analysis. T.S., X.X., N.Y. and I.I. assisted with data analysis and interpretation. N.Y. and I. I. supervised all the experiments. All authors contributed to the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tuvshintulga, B., Nugraha, A.B., Mizutani, T. et al. Development of a stable transgenic Theileria equi parasite expressing an enhanced green fluorescent protein/blasticidin S deaminase. Sci Rep 11, 9107 (2021). https://doi.org/10.1038/s41598-021-88594-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-88594-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.