Abstract
The increasing number of available genomic data allowed the development of phylogenomic analytical tools. Current methods compile information from single gene phylogenies, whether based on topologies or multiple sequence alignments. Generally, phylogenomic analyses elect gene families or genomic regions to construct phylogenomic trees. Here, we presented an alternative approach for Phylogenomics, named TOMM (Total Ortholog Median Matrix), to construct a representative phylogram composed by amino acid distance measures of all pairwise ortholog protein sequence pairs from desired species inside a group of organisms. The procedure is divided two main steps, (1) ortholog detection and (2) creation of a matrix with the median amino acid distance measures of all pairwise orthologous sequences. We tested this approach within three different group of organisms: Kinetoplastida protozoa, hematophagous Diptera vectors and Primates. Our approach was robust and efficacious to reconstruct the phylogenetic relationships for the three groups. Moreover, novel branch topologies could be achieved, providing insights about some phylogenetic relationships between some taxa.
Similar content being viewed by others
Introduction
Reconstruction of phylogenetic relationships has extensively been performed by molecular systematics; in which traditionally, different methods encompassing multiple sequence alignments and tree reconstruction algorithms analyze ribosomal sequences or conserved protein-coding genes1. Molecular phylogenetic trees are based on mutations differentially accumulated in orthologous gene pairs, being constructed either with DNA or amino acid sequences. Evolutionary changes in amino acid sequences are useful for long-term evolution information; because they are more conserved than DNA ones as they reflect the selection effects of non-synonymous nucleotide changes on codons2. However, choosing the right orthologous pair is not straightforward. Sequences that are very constrained are also very conserved, so no differences between species may be found. On the other hand, sequences that are very divergent can lead to distorted phylogenies.
The post-genomic era has provided large and unprecedent sequence datasets for thousands of organisms across several taxa of the Tree of Life. Consequently, molecular phylogenetics has benefited; phylogenomics has emerged as a relevant field, integrating molecular evolutionary analyses with genomic data3,4. Methods such as supertree5,6, supermatrices7,8,9, mega-phylogeny10 and multispecies coalescent model11,12 have been applied to reconstruct large phylogenies in a way that multi-gene phylogenies represent collectively a single evolutionary landscape.
Each method mentioned differs in some or many points among them, but all of them share the principle of combining individual gene phylogenies to plot a representative phylogenetic tree. Briefly, the supertree method relies on the compilation of topologies from several source gene trees for producing a single tree, whereas the supermatrix method is based on building a large multiple sequence alignment for simultaneous analyses of a giant phylogenetic matrix. Mega-phylogeny method is derivative from the latter, with some improvements during construction of multiple sequence alignments. Lastly, coalescent-based species tree method integrates population genetics processes with mathematical model to deal with heterogeneity and incongruity of gene trees to build a single tree.
Here, we present TOMM (Total Ortholog Median Matrix) as an alternative approach for phylogenomics, in which we propose the use of all orthologous pairs from the desired species for building a matrix based on their median amino acid distance obtained from the proteome (i.e., protein sequences of all protein-coding genes from a genome). Thus, we obtain a phylogeny based on the orthologous forest of sequences (an unsupervised strategy) rather than sets of trees knowingly selected (a supervised strategy).
TOMM retrieves orthologous proteins by using the Reciprocal Smallest Distance (RSD) method, which provides evolutionary distance measures used to build a distance matrix to obtain comprehensive phylograms. To evaluate the efficiency of such new approach, we have tested TOMM in three eukaryote groups of organisms: Kinetoplastida protist, Diptera hematophagous insects, and human and non-human Primates. We used these emblematic groups because of their relevance in the association among the taxa related to parasite-vector-host interaction. Moreover, this triad covers, in a modest way, a reasonable and feasible diversity of eukaryotes, including unicellular, invertebrate, and higher vertebrate organisms.
Kinetoplastid protists are flagellate excavates belonging to the phylum Euglenozoa. The members of the Kinetoplastea are characterized by the presence of circular DNA network disks (called kDNA) inside a large mitochondrion. This group presents a great biological variety, from free-living to parasitic organisms. Most known members belong to the family Trypanosomatidae, which are all obligate endoparasitic, comprising either monoxenous (single host, restricted to invertebrates) or dixenous (two hosts, a vertebrate or plant and an invertebrate vector) life cycles. The family Trypanosomatidae comprises 22 genera distributed in six formally recognized subfamilies13. Although most trypanosomatid genera are monoxenous, being able to infect only insects, this family is well known because of the dixenous genera Leishmania and Trypanosoma, which comprise species pathogenic to humans, causing serious insect-borne infectious diseases, such as leishmaniasis and Chagas’s disease, respectively. Because of the medically important species and their biological diversity, kinetoplastids represent an interesting model for understanding the evolution of both parasitism and pathogenicity.
The blood feeding habit evolved independently multiple times among the 400 hematophagous arthropod genera (over 14,000 species)14, including within the Diptera where it developed independently within the Brachycera (tsetse and tabanid flies), and at least twice in the suborder Nematocera to produce the mosquitoes and sand flies. These organisms are vectors of leishmaniasis, African trypanosomiasis, malaria, filariasis, and several viral diseases such as yellow fever, dengue, and zika.
Closing the triad, we performed the TOMM approach in higher vertebrates, represented herein by the Primates order, which is one of the most diverse among the mammals, comprising over 470 species15. Primates present extraordinary variations regarding ecological, behavioral, morphological, and evolutionary aspects. Genomic and genetic characterizations of primates are not only important for species conservation and evolutionary insights16,17, but also for understanding human evolution and genome structure from a biomedical perspective [reviewed in18]. Indeed, evolutionary genomics of host–pathogen interaction has been considered a trait for molecular phylogeny, and correlations between immunity against infections and Primates evolution have been targeted to understand how viral, bacterial, and parasitic diseases emerged to elucidate their different manifestations depending on host species19.
Overall, we implemented the TOMM phylogenomic approach for the three focal groups of organisms. The TOMM resulting trees are in good agreement with latest phylogenetic thoughts for the three groups of organisms.
Results and discussion
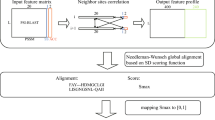
The overall procedure of TOMM approach is diagramed in Fig. 1. TOMM efficiently recovered known phylogenetic relationships and additionally was able to provide new phylogenetic insights. The three data sets analyzed herein produced well-resolved phylogenies. The Kinetoplastid tree (Fig. 2) showed congruent results with the most recent studies on this group13,20,21, with additional new possible relationships between some genera. Similarly, the hematophagous dipteran tree (Fig. 3) resembled the most recent phylogenetic relationships considered for the vectors of Malaria, viral diseases, leishmaniasis, and sleeping sickness22. For the Primates, TOMM phylogeny revealed two main clades, separating the most primitive primates (Strepsirrhini) from the other ones (Haplorrhini), that include Tasiiformes and Simiiformes. Among the haplorrhines, Platyrrhini formed a distinct well-supported clade from Catarrhini (Fig. 4), as expected18,23. However, TOMM was not efficient in recovering Cebus and Saimiri as a single clade of Cebidae family, clustering Cebus and Aotus in a non-supported clade (a.u. 55). Similarly, non-expected results were observed to C. atys and P. nubis, though with a high probability support (a.u. 98). The resulting trees are described and discussed in more detail hereafter.
Phylogenomic tree of Kinetoplastid protozoa. Phylogram constructed with the TOMM approach using approximately 5636 orthologous protein pairs across 46 Kinetoplastida species with genome sequence available (Table 1 and Supplemental Table S1). Numbers next to the branches represent the percentages of approximate unbiased support probabilities for 10,000 bootstraps, calculated using the pvclust package82 in R (R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2018, URL: https://www.R-project.org/). The Newick file was annotated using the program MEGA 6. Abbreviations: if (infra family); sg (subgenus); sf (subfamily).
Phylogenomic tree of hematophagous Diptera. Phylogram constructed with the TOMM approach using approximately 8168 orthologous protein pairs across 31 Diptera species with genome sequence available (Table 2 and Supplemental Table S2). Numbers next to the branches represent the percentages of approximate unbiased support probabilities for 10,000 bootstraps, calculated using the pvclust82 package in R (R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2018, URL: https://www.R-project.org/). The Newick file was annotated using the program MEGA 6. NW New World, OW Old World, GC gambiae complex.
Phylogenomic tree of Primates. Phylogram constructed with the TOMM approach using approximately 23,826 orthologous protein pairs across 25 Primates species with genome sequence available (Table 3 and Supplemental Table S3), and two outgroup species. Numbers next to the branches represent the percentages of approximate unbiased support probabilities for 10,000 bootstraps, calculated using the pvclust82 package in R (R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2018, URL: https://www.R-project.org/). The Newick file was annotated using the program MEGA 6.
Kinetoplastid tree
In the past decades, molecular phylogenetics using rRNA sequences and protein sequences have shed light in the evolutionary biology of this group, showing that parasitism evolved several times inside Kinetoplastea13,24. Comparative genomics of dixenous and monoxenous trypanosomatids were compared to the free-living kinetoplastid, Bodo saltans, thought to be the closest relative of the trypanosomatids13,20.
Important phylogenomics studies brought up key phylogenies across representative kinetoplastids from genera Leishmania, Trypanosoma, Phytomonas, Leptomonas, and Bodo20. More recently, such analyses were expanded to over 30 species encompassing several members for each life cycle: free-living, monoxenous, and dixenous parasites13. Collectively, these phylogenies were constructed using some gene families and a core of 64 conserved proteins. The TOMM approach has already been applied to Trypanosomatidae family in a study that identified a new parasite found in a fatal case of visceral leishmaniasis, where 36 genomes from trypanosomatids were used25. Here, we presented a phylogenomic analysis of kinetoplastids based on at least 1473 orthologous proteins across 45 species with published genomes, comprising flagellates of all lifestyles (see Table 1).
The resulting tables of pairwise orthologs were sorted to find the median value of the amino acid distance and thus populate a pairwise matrix (Supplemental Table 1, sheet “AA distance”). The minimum number of ortholog sequences found in the condition above was 1473, found for the Perkinsela sp./Phytomonas serpens pair, and the maximum was 8434 sequences, found for the Crithidia spp./Leptomonas pyrrhocoris proteome pair (Supplemental Table 1, sheet “Number-50”). This matrix was submitted to the program Pvclust, which provides statistical evaluation of the tree nodes expressed as approximately unbiased (a.u.) α values, where α = 1 − P. The phylogram was exported as a Newick file, including the a.u. values and annotated using the program MEGA 6.
The resulting phylogram built using the total proteome median matrix from kinetoplastid species harboring bacterial endosymbionts, free-living, monoxenous, and dixenous parasites is shown on Fig. 2. The enigmatic non-flagellated kinetoplastid Perkinsela spp, an obligate endosymbiont of Paramoeba (an amoeba genus considered an opportunistic pathogen of fish and marine invertebrates)26,27, is a clear outgroup. Usually, the free-living Bodo saltans, the closest known free-living relative of trypanosomatids, served as an outgroup for phylogenomics of trypanosomatids13,20. This Bodonidae species was placed in a sister position to Trypanosoma, whereas previous phylogenomic studies based on 64 well-conserved proteins strongly supported (1/100 BI posterior probabilities and ML bootstrap values) the late emergence of trypanosomatids as a sister group of Bodo saltans (Eubodonida)28. Here we were able to provide a higher statistical probability node support (a.u. 99), based on an average of 4999 orthologous proteins with a minimum (1833) and maximum (6022) ortholog pairs with Perkinsela spp and Trypanosoma theileri, respectively (Supplemental Table 1). In our analysis, between Trypanosoma spp. and B. saltans (free-living) is placed the monoxenous Paratrypanosoma confunsum, an early-branching trypanosomatid. In previous analyses, P. confusum branched at the base of the family Trypanosomatidae, representing a link between the ancestral free-living bodonids and the parasitic trypanosomatids13,21,29. The heterogeneity composition regarding the kinetoplastid life cycles make this clade a valuable source of information to elucidate the evolution of parasitism.
Corroborating the most recent expanded phylogeny tree of trypanosomatids from Lukes et al. (2018), the phylogram indicates the existence of two subfamilies with strong statistical support, Trypanosomatinae and Leishmaniinae. The Trypanosomatinae includes the parasites from the genus Trypanosoma, all dixenous species excepting T. evansi and T. equiperdum24, with important pathogens for humans and livestock. Trypanosoma vivax occupies a basal position within a clade with T. brucei, representing the African trypanosomes pathogenic to ungulates. The basal position of T. vivax in the clade is in accordance with previous results30,31.
The Leishmaniinae subfamily comprises two major Sections, Euleishmania (Leishmania) and Paraleishmania (Porcisia and Endotrypanum)32,33,34,35. The two sister clades, representing Euleishmania and Paraleishmania, were recently proposed as the infrafamily Leishmaniatae, whereas Crithidiatae infrafamily comprises all other genera of Leishmaniinae: Crithidia, Leptomonas, Lotmaria, Novymonas, Zelonia, and Borovskiya36,37.
The “Crithidiatae” clade is composed of monoxenous species and presented maximum statistical node support (a.u. 100), but subclades composed by Leptomonas, Lotmaria, and Crithidia species resulted from low node support (a.u. 56 and a.u. 51), with Crithidia fasciculata clustered apart from the Crithidia clade composed of C. bombi, C. expoeki, C. acanthocephali, and C. mellificae (a.u. 97). This reinforces the non-monophyletic origins of Crithidia members, and the revision of Crithidia genus as claimed by others38,39. Although monoxenous, Crithidiatae members, such as Leptomonas, have been detected in humans as co-infections in visceral leishmaniasis clinical cases40,41,42.
The Leishmaniatae (all dixenous) are clearly divided into the Leishmania and Viannia sub-genera, with L. tarentolae, a lizard parasite, occupying the most basal position in the Leishmania subgenus. Previously, this species was classified in the subgenus Sauroleishmania but was later shown from molecular phylogenetics to be closer to members of the Leishmania subgenus43,44. Leishmania enriettii from the subgenus Mundinia is located between the Viannia and Leishmania subgenera, as inferred by other phylogenetic studies45,46,47. However phylogenetic analyses, including other members of Mundinia, such as L. (Mundinia) martiniquensis and L. (M.) macropodum, support the most basal position of this subgenus in the genus Leishmania33,34,35. Endotrypanum is the only known kinetoplastid able to infect erythrocytes of their mammalian host (sloths)48. In the present study, E. monterogeii clearly clustered apart from all other subgenera of Leishmania as observed previously33,34,35. Between the clades Trypanosomatinae and Leishmaniinae, our analysis supports a clade sister to Leishmaniinae formed by two very-well supported clades: one comprising the genus Phytomonas (Phytomonadinae subfamily), whose species parasitize plants and another encompassing the bacterial-symbiont harboring genera, Strigomonas and Angomonas (Strigomonadinae subfamily). Interestingly, in our study Herpetomonas muscarum, which parasitize dipteran flies and Blechomonas ayalai, a parasite of fleas, formed a unique subclade sister to Strigomonadinae, a subfamily which includes bacterial endosymbiont harboring trypanosomatids of insects49,50,51,52. From these previous studies, the genus Herpetomonas is more closely related to the genera Phytomonas (transmitted to plants by phytophagous hemipterans) and Lafontella, the three genera forming the subfamily Phytomonadinae, whereas Blechomonas ayalai constituted the monogeneric blechomonadinae24,51. To the best of our knowledge, this is the first phylogenomic analysis that includes Herpetomonas and Blechomonas, whose species are found in closely related orders of insect hosts, Diptera and Siphonaptera, more phylogenetically related between them, than to Hemiptera, the order of the Phytomonas vectors53. However, phylogenomics, including more species of Herpetosoma, and the genus Lafontella, are still required to sustain this relationship.
To test the robustness of the method, we generated phylograms using the 25th and 75th percentiles instead of the median 50th percentile (Supplemental Figures S1 and S2, respectively), as well as running the RSD program with 1e−20 instead of 0.001 value for the blast including parameter, and 0.5 instead of 0.8 for the sequence length ratio including parameter (Supplemental Fig. S3). We also randomly reduced the proteomes to one half of their sizes and calculated the resulting median-based phylogram (Supplemental Fig. S4). They are all very similar, with some small deviations in the a.u. values, and T. vivax presented shifted placement within Trypanosoma cluster, but always with a.u. values smaller than 90.
Many orthology detection strategies are available, which raise many discussions about the ideal ortholog identification method, concerning to sensitivity and specificity. So far none of them is considered a gold standard54. The RSD method was applied within the TOMM pipeline because it is the only method which outputs an evolutionary distance measure. The OrthoMCL algorithm has been considered a balanced method identification and its database OrthoMCL-DB is a well-known portal for grouping orthologous protein sequences in a genome-scale across multiple species55. However, OrthoMCL does not provide an evolutionary distance measure. Regardless, we checked the RSD-derived orthologs with OrthoMCL-DB via TriTrypDB using the same set of species in which proteins were retrieved from the latter on (Table 1, as indicated in “Protein sequence source” column), in order to enable comparisons between the ortholog lists from obtained RSD and OrthoMCL-DB (Supplemental Table S4). From the total of 78 pairs of species comparison, an average of 87% ± 7.4% (Mean ± SD) of orthologs were detected by both methods. In half of the species combination (39 pairs), the RSD method was able to identify a higher number of orthologs in 16 pairs (20%), representing ortholog pairs exclusively detected by RSD over 50% higher than OrthoMCL-DB (pairwise comparisons with ratio of unique orthologs ≥ 1.5 at column “M” in Supplemental Table S4, e.g. L. enrietti vs T. evansi pair #38, which presented 755 unique orthologs with RSD against 446 unique orthologs with OrthoMCL). In turn, 29 pairs of species comparison (37%) represented number of orthologs exclusively detected by OrthoMCL-DB that were over 50% higher than RSD (e.g. pairwise comparisons with ratio of unique orthologs ≤ 0.5 at column “M” in Supplemental Table S4, e.g. L. tropica vs L. gerbilli pair #43, which presented 189 unique orthologs with OrthoMCL against 56 unique orthologs with RSD). In the remaining comparisons (43%) the number of unique orthologs detected by each method were homogenous (Supplemental Table S4). Overall, the orthology inference was very dependent for a given pair of species (e.g. Endotrypanum monterogeii vs Leishmania tarentolae or Leishmania gerbilli vs Trypanosoma evansi), but we observed few and homogenous differences between the number orthologs detected by OrthoMCL and RSD, without significant difference between them (Supplemental Fig. S7).
To further test the robustness of our approach, we employed another pipeline for identification of orthologs, using the SonicParanoid56 program with the MCL algorithm. This program produces an output with the predicted ortholog pairs from a two species comparison but lacks the calculation of the average amino acid distance between these pairs. We thus wrote a program that generated a fasta file containing the sequences of each ortholog pair, which was submitted to Clustal57 alignment, which in turn was submitted to a subroutine of the Mega X package58 to calculate the average amino acid distance for the pair. This allowed to generate a SonicParanoid-based amino acid distance matrix that was submitted to Pvclust as described above for the RSD-derived orthologs. The phylogenetic trees of kinetoplastid species generated by the TOMM-RSD and TOMM-SonicParanoid methods can be viewed in Supplemental Figs. S5 and S6. All the branches of the trees depicting the various subgroups are congruent. The main difference between the trees is the location of Boldo saltans, which is within the Trypanosomatidae in the RSD-derived tree, with a support of 92%, but in the SonicParanoid tree it is located in between the Leishmanidae and Trypanosomatidae. We conclude that the use of an alternate method of determining the orthologs does not affect the results of the TOMM approach to phylogeny determination. The Sonic approach has the advantage of being very fast compared with the RSD, but the lack of an output of the paired amino acid distances removes this advantage compared to the RSD method. It would be very useful if the sonic paranoid pipeline included the resulting average amino acid distance of the ortholog pairs.
Hematophagous dipteran tree
The phylogenomic tree for Diptera vectors was built with 29 species from Brachycera (Tsetse flies, Glossina) and Nematocera (the majority are Anopheles mosquitoes) suborders (Table 2), using the non-hematophagous D. melanogaster as outgroup and M. domestica as a comparator species for Glossina genus. Here, the main vectors related to Kinetoplastid parasites are species from the Glossina genus and Psychodidae family (sandflies), which transmit, respectively, African Trypanosoma and Leishmania protozoans. Hematophagous hemipterans from the subfamily Triatominae are another important group of vectors for Trypanosoma parasites; however they were not considered here, because of the high distance in phylogenetic relationship between the Diptera and Hemiptera orders. In fact, due to the great diversity of insects, even inside the Diptera order, it is observed as a very large distance among the families. Such diversity can be verified by the wide range in genome sizes and number of protein-coding genes shown in Table 2.
The phylogram for hematophagous dipterans was based on an average of 8168 orthologous proteins, with a minimum number of ortholog sequences (5893) found in Anopheles maculatus/ Lutzomyia longipalpis pair of vectors species. The highest number of2 ortholog sequences was 13,161 between the Tsetse flies Glossina austeni and Glossina pallipides (Supplemental Table ). To the best of our knowledge, these are the highest numbers of orthologous genes considered for taxa inside Diptera, as collectively surveyed previously59.
The well-known D. melanogaster was considered an outgroup species to hematophagous dipterans, but as a dipteran it has not presented a proper isolation of an outgroup, being positioned inside the highly supported Brachycera clade (a.u. 100). In general, as observed for Primates and Kinetoplastida, the TOMM approach was also robust in building the phylogenetic relationships to this group of dipterans (Fig. 3). The Nematocera clade presented a moderate support (a.u. 73), which can be explained by a split in two families, Psychodidae (Lutzomyia longipalpis and Phlebotomus papatasi) and Culicidae (Anopheles, Aedes and Culex) (Fig. 3). Interestingly, in previous insect phylogenomics studies, Culicidae species have been placed apart from all other dipterans, and although more externally, Psychodidae is positioned in the same clade with Glossina and Drosophila54. However, here we have found an opposite topology reached by TOMM phylogram for Psychodidae species, in which Phlebotomus and Lutzomyia were more closely related to Culicidae (all Nematocera) than the Brachycera species (Glossina genus).
The evolutionary relationships of Anopheline mosquitoes are widely studied because of the great medical importance of this group as vectors of Malaria, especially the Anopheles gambiae complex, which is composed of eight species morphologically indistinguishable; however the species display differential traits such as, behavior, ecological niche, and vector competence60. Using whole-genome reference sequences, different phylogenetic relationships between genomic regions have been inferred for A. gambiae complex when differential analyses target autosomes or sex chromosomes and coding or non-coding loci60,61. A consensus phylogenetic relationship between A. gambiae (G) and A. coluzzi (C) as a sister group (G + C) was found in two comprehensive studies using X chromosome or autosomes, employing Maximum-Likelihood- (ML)60 or Bayesian Multispecies Coalescent model-61 based methods. In addition, another sister group composed of A. arabiensis (A) and A. quadrianulatus (Q) was inferred only when X chromosome genomic regions were used60,61.
Here, the clade topology of A. gambiae complex reached by TOMM approach (Fig. 3) corroborates the sister group A + Q inferred by known X chromosome phylogenies with high confidence (a.u. 98). However, the topology for other species relationships depicted a different scenario. Of note, G + C were not placed together in a same branch and A. merus (R), often branched in a more external position of the trees, was significantly (a.u. 96) placed more internally close to A + Q pair. Moreover, A. melas (L) was the earliest branched species in the clade; whereas in known phylogenies, A. merus was placed in this position. Thus, while the most recent topologies61 for A. gambiae complex presented patterns as (R((L(A + Q))(G + C))) for non-coding and ((L(A + Q))(R(G + C))) for coding data from X chromosome, the TOMM approach reassembled the pattern (L(G(C(R(A + Q))))) using all sets of orthologous proteins (over 8000 coding sequences) found for the 29 species used.
Primates tree
The Primates phylogenomic tree included 25 species presenting published whole-genome sequence, encompassing all sublevels of the order, including lemurs, lorises, tarsiers, New World Monkeys (NWM), Old World Monkeys (OWM), big apes, and humans15, and includes the two additional mammals species that were used as outgroups (see Table 3). The Primates phylogram was based on aveage 23,826 orthologous proteins, with a minimum number of ortholog sequences (19,185) found in Propithecus coquereli/Carlito syrichta pair of primate species. If considered the entire phylogram, which includes the two outgroups species, the overall minimum number of ortholog sequences was 18,970 (Tupaia chinensis/Carlito syrichta pair). The highest number of ortholog sequences was 39,341 between Homo sapiens and Pan troglodytes, showing that the topology achieved by the TOMM approach accounts for both the number of orthologs, as well as amino acid distances (Supplemental Table 3).
The Primates phylogram showed correctly Mus musculus as an outgroup and several well-formed clades within the Strepsirrhini and Haplorrhini suborders (Fig. 4). Main taxonomic groups at suborder sublevels (Catarrhini and Platyrrhini), as well as at family level (Cercopithecidae and Hominidae), resemble current knowledge (Lockwood et al. 2004; Langergraber et al. 2012; Freitas et al. 2018). Among the superfamily Hominoidae, the human location and its relationship with the gorilla and chimpanzee/bonobo clades (a.u. 100) from the Homininae subfamily was similar to that shown in previous studies62,63,64, suggesting an accelerated evolution of human genes, as proposed by Hubisz and Pollard65. The position of Nomascus leucogenys, the critically endangered gibbon from the Hylobatidae family, is also accurate66. However, two clades showed different clustering compared to other Primates phylogenomic studies18,23: one regarding OWM (Catarrhini) from Cercopithecoidea (highlighted in blue) and another clustering NWM (Plathyrrhini) from Aotus and Cebus genres (highlighted in red) (Fig. 4).
Cercocebus atys is an OWM, who inhabits the West African forests (from Senegal and Congo), considered, by IUCN, as Vulnerable (VU)66. This species is naturally infected by the Simian Immunodeficiency Virus (SIVsmm), and due to its close-relationship with humans, the hazardous form of this virus, HIV-2 (Human Immunodeficiency Virus, type 2), was transmitted to man67. Such genus has been commonly placed closer to the baboons from Mandrillus genus68,69. However, we did not use any protein collection from Mandrillus species in our approach. The most related species from Papionini tribe used herein was from Macaca genus and from the widest-ranging baboon Papio anubis, which clustered with C. atys, and then to Macaca species, that showed highly supported clades.
Related to the NWM platyrrines, Cebus capucinus from the Cebidae family clustered with the only night monkey species with complete genome sequence available, Aotus nancymaae from the Aotidae family, rather than the other Cebidae representative, Saimiri boliviensis (Fig. 4). Aotus neotropical monkeys are often used as a primate biological model for Plasmodium infection in Malaria researches70, raising extensive discussions about their evolutionary relationships with other NWM71. Classical overviews on adaptive radiation of neotropical primates, discussing phylogenetic relationships and inconsistences among Saimiri, Cebus and Aotus, highlighted discordances between morphological and molecular analyses72,73. Nevertheless, mostly molecular approaches have usually considered Saimiri and Cebus as representatives from the Cebidae family, and Aotus as a distinct clade from Aotidae63,64,73. Such results were also observed by the most complete primate mitogenomics performed to date17. Our TOMM phylogenomic tree revealed a low probability supported clade (a.u. 55), clustering Aotus and Cebus when a cutoff value of 50% was considered. Such unresolved clustering may have been shaped by influence of the total number of orthologous proteins found among the three species, since Aotus-Cebus pair presented more orthologous proteins (25,629), than Saimiri-Cebus (24,085) or Aotus-Saimiri (23,205) (Supplemental Table 3). Thus, the results presented here should maintain this evolutionary debate within the field of primatology.
Concluding remarks
Even with genomic data available for several groups of organisms along the tree of life, reaching a definitive evolutionary relationship among taxa is still hard. That is because evolution of genomes undergoes great dynamic evolutionary processes with different pressures depending on the genomic region and gene product function. Evaluating phylogenomic relationships depends on numerous supervised methods and procedures, all subject to variable benefits and disadvantages, where a trade-off between accuracy and objectivity is pondered relying on the type of application. Despite all these caveats, there is no hesitation that Phylogenomics is a powerful integrated field that is raising key questions in the evolutionary history of several group of organisms and providing very useful information, whether for biodiversity conservation or in agriculture, livestock, and biomedical matters.
Here, we presented the TOMM approach for phylogenomic analysis, which uses genome-wide protein-coding sequences for a given group of organisms, gathering orthologous predicted proteomes between pairs of desired taxa in order to build a single phylogram based on their median amino acid evolutionary distances. This unsupervised approach was basically divided in two extensive steps, where the first consists of orthology inference and the second is composed of steps to build a large pairwise amino acid distance matrix; this latter is the novelty along the rational analysis for Phylogenomics.
Regarding the first step, as any other phylogenetic analysis, TOMM approach relies on inferring orthologs. Reliable orthologs identification between genome sequences is challenged by how different evolutionary mechanisms operate in different genomic regions. As surveyed and discussed elsewhere74, there are several methods for orthology inference, all presenting advantages and limitations, but the most common methods are based on sequence similarity. Here, we used Reciprocal Smallest Distance (RSD) method75, which is obtained from sequence similarity metrics within an evolutionary distance matrix. Also, RSD uniquely provides an amino acid distance measure. Many different orthology inference methods were not evaluated during TOMM approach, because our aim was not to test orthology detection performance, rather to perform a comprehensive phylogenomic analysis based on all pairwise orthologous pairs found inside a group.
Since there is no choice of gene families or genomic regions, as many phylogenomic studies ascribed them, we denominate our approach as unsupervised and “total”. The originality of our phylogenomic analysis is related to the second step of procedure, through the construction of a species matrix populated with evolutionary distance measurements calculated in the previous step, rather than performing multiple sequence alignments. However, sequence alignments were embedded during orthology detection. We assigned the “median” amino acid distance between two taxa as a measurement to populate the species matrix and then building the phylogram, but by testing other percentiles of distance measures, we observed that the TOMM approach has kept the robustness of results about well-known phylogenetic relationships.
Possible criticisms concerned to our approach are i) the computational resources needed, because the RSD method is computationally intensive, and it worsens for large genomes and ii) the customized programs to help building the amino acid distance matrix are operational system-restricted (Windows Microsoft). The first step is not feasible to common PC machines and it must be performed within HPC resources. However, with the increasing availability of HPCs whether offered by public or private institutions and virtual machines as emulators of computational systems, make these two concerns minor caveats. Another concern is related to sampling taxa; the benefit of use the total predicted proteomes has a limitation in the number of publicly available organisms with annotated genome sequences. Even though, we showed here that TOMM approach is applicable and robust for wide range of taxa presenting distinct genome sizes and complexity, since we applied to Kinetoplastid (9.5–58.7 Mb haploid genome size), hematophagous Diptera (137–1923 Mb haploid genome size), and Primates (2487–3454 Mb haploid genome size). Its robustness was also verified when trees were generated from genomes reduced randomly to 50% of their sizes, when very similar trees were obtained (Supplemental Fig. S4).
Finally, this approach was not only able to corroborate the main knowledge in phylogenetic relationships of tested groups of organisms, but also to present novel branch topologies. We believe that our results with TOMM should contribute to supporting and enriching the evolutionary insights to the field.
Methods
Sequence datasets
We used protein sequences of all protein-coding genes (proteome), deduced from a complete genome for a given species, downloading data from Kinetoplasdida, Diptera, and Primates, as well as other external organisms (Table 1, 2, 3).
The Kinetoplastid genomic sequences from 46 species were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome) or TriTrypDB (http://tritrypdb.org/tritrypdb/) databases, according to information provided in Table 1. The protein sequences corresponding to coding sequences from a given Kinetoplastid genome were downloaded when available. When protein sequences were not available, as the genes of these organisms do not contain introns, we straightforwardly translated them in-house from genomic sequences by obtaining open reading frames from six the translations using the EMBOSS tool76 and adjusting the starting Methionine by BLASTX to the Protozoa-RefSeq NCBI database. This information was specified in Table 1 at “protein sequence source” column. Perkinsela sp. was used as outgroup.
For the hematophagous dipterans dataset, all protein sequences were downloaded from VectorBase77 (https://www.vectorbase.org/downloads) as specified in Table 2, except for Drosophila melanogaster, which was downloaded from RefSeq NCBI database. Both the non-hematophagous flies, Musca domestica and D. melanogaster. were used as related species.
For the Primates, we used annotated complete genomes of 25 species, including Homo sapiens. Mus musculus (House mouse) and Tupaia chinensis (Chinese tree shrew) were used as an outgroup. All protein sequences of the mammalians were downloaded from RefSeq Protein NCBI database (https://www.ncbi.nlm.nih.gov/refseq/) (Table 3).
Data analyses
The TOMM pipeline was performed in several steps, as shown in Fig. 1. First, the protein sequences were dereplicated, and then clustered at 99% identity. The centroids were saved using the Usearch program version 9.078. Only downloaded protein sequences or translated protein-coding genomic sequences larger than 50 amino acids were used in the subsequent analyses. To sample the proteome to 50% of its level, we used the program Seqtk available at https://github.com/lh3/seqtk. Second, the proteomes from each of the downloaded genomes (or translated coding sequences in-house) were pairwise submitted to the program Reciprocal Smallest Distance (RSD)75 to obtain a table of orthologs and their amino acid (aa) distances. The RSD algorithm employs global sequence alignment by using ClustalW79 and maximum likelihood by using PAML80 to estimate the amino acid substitutions. To build the matrix of median pairwise amino acid distances (AAD) from genome-derived protein sequences, pairs of proteomes [the number of pairs is equal to (n2 – n)/2, where n = number of species], for each taxonomic group used here, were submitted to the program RSD using the NIH Biowulf cluster (https://hpc.nih.gov/systems/). For the Kinetoplastida and hematophagous Diptera, we used the RSD settings of 0.001 for the blast e-value of acceptance, and the value of 0.8 for the minimum ratio of the smallest sequence to the larger one. For Primates, the e-value of acceptance was 0.1. The RSD tables were sorted by their AAD’s to obtain the desired percentile values of AAD. Matrices were constructed for specified percentile values. These matrices were then submitted to the Hclust81 and Pvclust82 packages into R version 3.5.283 to obtain phylograms, after 10,000 bootstraps. The APE package84 was used to export the trees (in Newick format), and these were annotated using the MEGA 6 software85. The approximately unbiased values of the nodes (expressed as α values, where α = 1- P), as provided by Pvclust, were exported to a Newick file by modifying a function provided at https://stackoverflow.com/questions/22749634/how-to-append-bootstrapped-values-of-clusters-tree-nodes-in-newick-format-in. The R script for these operations is shown in Supplemental File 1.
To compare the orthologs identified by RSD with those inferred by MCL algorithm, we used SonicParanoid. A in-house script compiled the protein sequences of each ortholog pair in a fasta file, which in turn was submitted to multiple sequence alignment (MSA) using Clustal57. Then, the amino acid divergence was calculated using the MSA in a routine of the the Mega X package58, resulting in a SonicParanoid-based amino acid distance matrix.
To compare the orthologs detected by RSD with those of the TriTypDB database, searches were performed using the TriTypDB database (https://tritrypdb.org). For this, all genes of the species Endotrypanum monterogeii, Leishmania aethiopica, Leishmania arabica, Leishmania enriettii, Leishmania tropica, Leishmania gerbilli, Leishmania turanica, Leishmania tarentolae, Trypanosoma evansi, Trypanosoma vivax, Trypanosoma theileri, Blechomonas ayalai and Crithidia fasciculata were compared in pairs using “Identify Genes based on Orthology Phylogenetic Profile” tool, determined by the OrthoMCL algorithm86 under OrthoMCL-DB, yielding 78 pairwise comparisons. For method comparison, intersections between RSD-derived and OrthoMCL-derived orthologs were calculated using respective gene ID lists as input in custom Venn diagram tool available at http://bioinformatics.psb.ugent.be/webtools/Venn/.
Customized In-House programs to retrieve orthologous sequence from RSD
Three programs were written in Visual Basic v6.0 to facilitate the step of orthologous identification in the pipeline. These are named RSD-maker, Batcher, and RSD2Table. They are available for download at https://s3.amazonaws.com/proj-bip-prod-publicread/transcriptome/Tomm/Tomm-executables.zip.
RSD-maker takes as input a list of FASTA file names and produces a tab-delimited list of all pairs of FASTA files to be submitted to RSD. It can take also an additional list of FASTA pairs already processed, and in this case, it outputs only the missing pairs. This is useful when an additional proteome is added after RSD has been run on a group of sequences. The sequence pairs for each pairwise RSD comparison are then provided as input to the program Batcher, which also takes as input the command line for the RSD program, such as “rsd_search -q INPUT1 -subject-genome = INPUT2 -outfmt 1 -de 0.8 0.1 -o output/INPUT1-INPUT2-0.8_0.1.tbl”. Upon running the program, INPUT1 and INPUT2 will be substituted by the tab-delimited pair to produce a file containing hundreds or thousands of commands as dictated by the number of pairs used as input (RSD resulting files). Such resulting file is used to run simultaneously as a swarm in the NIH Biowulf HPC (High-Performance Computing; http://hpc.nih.gov). The RSD resulting files (Supplemental File 2, as compressed folders “RSD-Primates”, “RSD-Flies”, RSD-kinetoplastids”) contain gene ID lists tabulated for INPUT1 species (first column) and INPUT2 (second column), they are then processed by the program RSD2Table. It takes as input the list of FASTA files as well as a list of the RSD results, and the desired percentile value. It then sorts the RSD files in ascending order of the AAD values and finds the AAD corresponding to the desired percentile. This program can also receive a list of desired percentiles and then produces all matrices in a single run. In addition to the aa distance matrix of the orthologs, it also produces a table indicating the number of ortholog pairs found by RSD. The matrices are written as “table-10.tbl” or “table-50.tbl”, where 10 and 50 are the pre-determined percentiles. These matrices can then be submitted to the program Batcher, that will take as INPUT1 the list of percentiles and the R script shown in Supplemental File 1, to produce an output that can be pasted on the R console to produce the Pvclust results and Newick file as described in the previous paragraph.
The main computationally intensive job for identification of orthologous sequences is the calculation of the RSDs, which can take a few hours per CPU for the smaller Kinetoplastid genomes, to over one day for the larger genomes such as from the Primates. For example, the 27 mammal species used in this work lead to 351 pairwise comparisons, which could consume over one year of computational time for a single CPU. However, no more than 4 GB of memory is needed per CPU, and the job can be easily parallelized on an HPC system, so the results were obtained in approximately two days.
Data availability
The Supplemental File 2 is a compressed folder available in the Dryad repository at https://doi.org/10.5061/dryad.b1k526g, as well as all other supplemental data supporting the results of this article.
References
Pyron, R. A. Post-molecular systematics and the future of phylogenetics. Trends Ecol. Evol. 30, 384–389 (2015).
Nei, M. & Kumar, S. Molecular Evolution and Phylogenetics (Oxford University Press, Oxford, 2000).
Eisen, J. A. & Fraser, C. M. Phylogenomics: intersection of evolution and genomics. Science 300, 1706–1707 (2003).
Delsuc, F., Brinkmann, H. & Philippe, H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 6, 361–375 (2005).
Bininda-Emonds, O. R. P. The evolution of supertrees. Trends Ecol. Evol. (Amst.) 19, 315–322 (2004).
Moore, B. R., Smith, S. A. & Donoghue, M. J. Increasing data transparency and estimating phylogenetic uncertainty in supertrees: approaches using nonparametric bootstrapping. Syst. Biol. 55, 662–676 (2006).
McMahon, M. M., Sanderson, M. J. & Savolainan, V. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst. Biol. 55, 818–836 (2006).
de Queiroz, A. & Gatesy, J. The supermatrix approach to systematics. Trends Ecol. Evol. 22, 34–41 (2007).
Philippe, H. et al. Pitfalls in supermatrix phylogenomics. Eur. J. Taxonomy 283, 1–25 (2017).
Smith, S. A., Beaulieu, J. M. & Donoghue, M. J. Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol. Biol. 9, 37 (2009).
Liu, L., Wu, S. & Yu, L. Coalescent methods for estimating species trees from phylogenomic data - Liu - 2015 - Journal of Systematics and Evolution - Wiley Online Library. J. Syst. Evol. 53, 380–390 (2015).
Edwards, S. V. et al. Implementing and testing the multispecies coalescent model: a valuable paradigm for phylogenomics. Mol. Phylogenet. Evol. 94, 447–462 (2016).
Lukeš, J. et al. Trypanosomatids are much more than just trypanosomes: clues from the expanded family tree. Trends Parasitol. 34, 466–480 (2018).
Ribeiro, J. M. Blood-feeding arthropods: live syringes or invertebrate pharmacologists?. Infect. Agents Dis. 4, 143–152 (1995).
Mittermeier, R. A., Rylands, Anthony B. & Wilson, D. E. Handbook of the Mammals of the World. Volume 3 - Primates. vol. 3 (Lynx Edicions, 2013).
Ayala-Burbano, P. A. et al. Genetic assessment for the endangered black lion tamarin Leontopithecus chrysopygus (Mikan, 1823), Callitrichidae, Primates. Am. J. Primatol. 79, e22719 (2017).
de Freitas, P. D. et al. Next-generation sequencing of the complete mitochondrial genome of the endangered species black lion Tamarin Leontopithecus chrysopygus (Primates) and Mitogenomic Phylogeny focusing on the callitrichidae family. G3 Genes Genomes Genet. 8, 1985–1991 (2018).
Pecon-Slattery, J. Recent advances in primate phylogenomics. Annu. Rev. Anim. Biosci. 2, 41–63 (2014).
Brinkworth, J. F. & Pechenkina, K. Primates, Pathogens and Evolution (Springer, New York Heidelberg Dordrecht London Library of, 2013).
Jackson, A. P. et al. Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Curr. Biol. 26, 161–172 (2016).
Skalický, T. et al. Extensive flagellar remodeling during the complex life cycle of Paratrypanosoma, an early-branching trypanosomatid. PNAS 114, 11757–11762 (2017).
Neafsey, D. E. et al. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347, 1258522 (2015).
Springer, M. S. et al. Macroevolutionary dynamics and historical biogeography of primate diversification inferred from a species supermatrix. PLoS ONE 7, e49521 (2012).
Maslov, D. A. et al. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology 146, 1–27 (2019).
Maruyama, S. R. et al. Non-leishmania parasite in fatal visceral leishmaniasis–like disease, Brazil. Emerg. Infect. Dis. J. https://doi.org/10.3201/eid2511.181548 (2019).
David, V. et al. Gene loss and error-prone RNA editing in the mitochondrion of perkinsela, an endosymbiotic kinetoplastid. mBio 6, e01498-15 (2015).
Tanifuji, G. et al. Genome sequencing reveals metabolic and cellular interdependence in an amoeba-kinetoplastid symbiosis. Sci. Rep. 7, 11688 (2017).
Deschamps, P. et al. Phylogenomic analysis of kinetoplastids supports that trypanosomatids arose from within bodonids. Mol. Biol. Evol. 28, 53–58 (2011).
Flegontov, P. et al. Paratrypanosoma is a novel early-branching trypanosomatid. Curr. Biol. 23, 1787–1793 (2013).
Jackson, A. P. et al. A cell-surface phylome for African trypanosomes. PLOS Neglect. Trop. Dis. 7, e2121 (2013).
Jackson, A. P. et al. Global gene expression profiling through the complete life cycle of trypanosoma vivax. PLOS Neglect. Trop. Dis. 9, e0003975 (2015).
Cupolillo, E., Medina-Acosta, E., Noyes, H., Momen, H. & Grimaldi, G. A revised classification for leishmania and endotrypanum. Parasitol. Today 16, 142–144 (2000).
Espinosa, O. A., Serrano, M. G., Camargo, E. P., Teixeira, M. M. G. & Shaw, J. J. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 145, 430–442 (2016).
Barratt, J. et al. Isolation of novel Trypanosomatid, Zelonia australiensis sp. nov. (Kinetoplastida: Trypanosomatidae) provides support for a gondwanan origin of dixenous parasitism in the Leishmaniinae. PLOS Negl. Trop. Dis. 11, e0005215 (2017).
Kaufer, A., Barratt, J., Stark, D. & Ellis, J. The complete coding region of the maxicircle as a superior phylogenetic marker for exploring evolutionary relationships between members of the Leishmaniinae. Infect. Genet. Evol. 70, 90–100 (2019).
Jirků, M., Yurchenko, V. Y., Lukeš, J. & Maslov, D. A. New species of insect trypanosomatids from costa rica and the proposal for a new subfamily within the trypanosomatidae. J. Eukaryot. Microbiol. 59, 537–547 (2012).
Kostygov, A. Y. & Yurchenko, V. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae). Folia Parasitol. 64, 020 (2017).
Hollar, L., Lukeš, J. & Maslov, D. A. Monophyly of endosymbiont containing trypanosomatids: phylogeny versus taxonomy. J. Eukaryot. Microbiol. 45, 293–297 (2007).
Yurchenko, V. Y., Lukeš, J., Tesařová, M., Jirků, M. & Maslov, D. A. Morphological discordance of the new trypanosomatid species phylogenetically associated with the genus crithidia. Protist 159, 99–114 (2008).
Ghosh, S., Banerjee, P., Sarkar, A., Datta, S. & Chatterjee, M. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian Leishmaniasis. J. Clin. Microbiol. 50, 2774–2778 (2012).
Singh, N., Chikara, S. & Sundar, S. SOLiDTM sequencing of genomes of clinical isolates of Leishmania donovani from India confirm leptomonas co-infection and raise some key questions. PLoS ONE 8, e55738 (2013).
Selvapandiyan, A., Ahuja, K., Puri, N. & Krishnan, A. Implications of co-infection of Leptomonas in visceral leishmaniasis in India. Parasitology 142, 1657–1662 (2015).
Briones, M. R. et al. Leishmania tarentolae taxonomic relatedness inferred from phylogenetic analysis of the small subunit ribosomal RNA gene. Mol. Biochem. Parasitol. 53, 121–127 (1992).
Croan, D. G., Morrison, D. A. & Ellis, J. T. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol. Biochem. Parasitol. 89, 149–159 (1997).
Puechberty, J. et al. Compared genomics of the strand switch region of Leishmania chromosome 1 reveal a novel genus-specific gene and conserved structural features and sequence motifs. BMC Genom. 8, 57 (2007).
Leelayoova, S. et al. Multilocus characterization and phylogenetic analysis of Leishmania siamensis isolated from autochthonous visceral leishmaniasis cases, southern Thailand. BMC Microbiol. 13, 60 (2013).
Ludwig, A. & Krieger, M. A. Genomic and phylogenetic evidence of VIPER retrotransposon domestication in trypanosomatids. Mem. Inst. Oswaldo Cruz 111, 765–769 (2016).
Franco, A. M. R. & Grimaldi, G. Jr. Characterization of endotrypanum (Kinetoplastida: Trypanosomatidae), a unique parasite infecting the neotropical tree sloths (Edentata). Memórias do Instituto Oswaldo Cruz 94, 261–268 (1999).
Alves, J. M. P. et al. Genome evolution and phylogenomic analysis of candidatus kinetoplastibacterium, the betaproteobacterial endosymbionts of strigomonas and angomonas. Genome Biol. Evol. 5, 338–350 (2013).
Borghesan, T. C. et al. Molecular phylogenetic redefinition of herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist 164, 129–152 (2013).
Votýpka, J. et al. Diversity of Trypanosomatids (Kinetoplastea: Trypanosomatidae) parasitizing fleas (Insecta: Siphonaptera) and description of a new genus Blechomonas gen. n. Protist 164, 763–781 (2013).
Votýpka, J. et al. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist 165, 825–838 (2014).
Misof, B. et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767 (2014).
Chen, F., Mackey, A. J., Vermunt, J. K. & Roos, D. S. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2, e383 (2007).
Chen, F., Mackey, A. J., Stoeckert, C. J. & Roos, D. S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34, D363–D368 (2006).
Cosentino, S. & Iwasaki, W. SonicParanoid: fast, accurate and easy orthology inference. Bioinformatics 35, 149–151 (2019).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Behura, S. K. Insect phylogenomics. Insect Mol. Biol. 24, 403–411 (2015).
Fontaine, M. C. et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347, 1258524 (2015).
Thawornwattana, Y., Dalquen, D. & Yang, Z. Coalescent analysis of phylogenomic data confidently resolves the species relationships in the anopheles gambiae species complex. Mol. Biol. Evol. 35, 2512–2527 (2018).
Olson, M. V. & Varki, A. Sequencing the chimpanzee genome: insights into human evolution and disease. Nat. Rev. Genet. 4, 20–28 (2003).
Lockwood, C. A., Kimbel, W. H. & Lynch, J. M. Morphometrics and hominoid phylogeny: support for a chimpanzee-human clade and differentiation among great ape subspecies. Proc. Natl. Acad. Sci. U.S.A. 101, 4356–4360 (2004).
Langergraber, K. E. et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 15716–15721 (2012).
Hubisz, M. J. & Pollard, K. S. Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr. Opin. Genet. Dev. 29, 15–21 (2014).
IUCN. The International Union for Conservation of Nature’s Red List of Threatened Species. (2019).
Ling, B. et al. Classic AIDS in a sooty mangabey after an 18-year natural infection. J. Virol. 78, 8902–8908 (2004).
Page, S. L., Chiu, C. & Goodman, M. Molecular phylogeny of old world monkeys (Cercopithecidae) as inferred from γ-Globin DNA sequences. Mol. Phylogenet. Evol. 13, 348–359 (1999).
Liedigk, R., Roos, C., Brameier, M. & Zinner, D. Mitogenomics of the old world monkey tribe papionini. BMC Evol. Biol. 14, 176 (2014).
Herrera, S., Perlaza, B. L., Bonelo, A. & Arévalo-Herrera, M. Aotus monkeys: their great value for anti-malaria vaccines and drug testing. Int. J. Parasitol. 32, 1625–1635 (2002).
Schneider, H. & Sampaio, I. The systematics and evolution of New World primates—a review. Mol. Phylogenet. Evol. 82, 348–357 (2015).
Schneider, H. & Rosenberger, A. Molecules, morphology, and Platyrrhine systematics. In Adaptive Radiations of Neotropical Primates 3–19 (Springer US, 1996).
Osterholz, M., Walter, L. & Roos, C. Retropositional events consolidate the branching order among New World monkey genera. Mol. Phylogenet. Evol. 50, 507–513 (2009).
Tekaia, F. Inferring orthologs: open questions and perspectives. Genomics Insights 9, GEI.S37925 (2016).
Wall, D. P. & Deluca, T. Ortholog detection using the reciprocal smallest distance algorithm. Methods Mol. Biol. 396, 95–110 (2007).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277 (2000).
Giraldo-Calderón, G. I. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43, D707–D713 (2015).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics 13, 555–556 (1997).
Mullner, D. fastcluster: fast hierarchical, agglomerative clustering routines for R and Python. J. Stat. Softw. 53, 1–18 (2013).
Suzuki, R. & Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006).
R Core Team. R: A language and environment for statistical computing. http://www.R-project.org/. (2018).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Li, L., Stoeckert, C. J. & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Acknowledgements
The authors thank Brian Brown, NIH Library Writing Center, for manuscript editing assistance. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Funding
This work was supported by São Paulo Research Foundation (FAPESP, Young Investigator Award, Grant 2016/20258-0 to SRM). SRM received a fellowship from FAPESP (2017/16328-6). LAR received a scholarship from FAPESP (2018/26799-9). JMCR was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Contributions
J.M.R. conceived and developed the methodological procedure. S.R.M., L.A.R., P.D.F., M.M.G.T. and J.M.R. performed the data analyses. S.R.M. and J.M.R. drafted the manuscript. S.R.M. wrote the final version of manuscript. LAR, PDF, MMGT and JMR proofread the manuscript. SRM and MMGT interpreted and discussed the results for Kinetoplastida organisms; S.R.M. and P.D.F. interpreted and discussed the results for Primates organisms; S.R.M. and J.M.R. interpreted and discussed the results for hemataphagous Diptera organisms. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maruyama, S.R., Rogerio, L.A., Freitas, P.D. et al. Total Ortholog Median Matrix as an alternative unsupervised approach for phylogenomics based on evolutionary distance between protein coding genes. Sci Rep 11, 3791 (2021). https://doi.org/10.1038/s41598-021-81926-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-81926-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.