Abstract
The importance of dynamic factors in enzyme evolution is gaining recognition. Here we study how the evolution of a new enzymatic activity exploits conformational tinkering and demonstrate that conversion of a dimeric phosphotriesterase to an arylesterase in Pseudomonas diminuta is accompanied by structural divergence between the two subunits. Deviations in loop conformations increase with promiscuity, leading to functionally distinct states, while they decrease during specialisation for the new function. We show that opposite loop movements in the two subunits are due to a dynamic coupling with the dimer interface, the importance of which is also corroborated by the co-evolution of the loop and interface residues. These results illuminate how protein dynamics promotes conformational heterogeneity in a dimeric enzyme, leading to alternative evolutionary pathways for the emergence of a new function.
Similar content being viewed by others
Introduction
Proteins are dynamic entities, with a wide spectrum of conformational rearrangements linked to their functions1,2. Protein motions are crucial to optimise substrate interactions3,4,5, but their role in enzymatic catalysis is highly debated6,7. Here we were interested in the role of protein dynamics in the emergence of new enzymatic functions.
Evolution of enzymes usually involves multifunctional intermediates8, indicating that the active site can accommodate more than one substrate9. Increasing experimental data demonstrates that enzyme multi-functionality is a widespread phenomenon8,10. Thus, new functions can emerge, without considerable cost to the original activity11. Different active site geometries are optimized for alternative catalytic strategies12 or carry out distinct activities13,14. Specialisation for different activities also depends on the network context10, which modulates the populations of different conformational sub-states15.
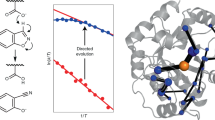
Protein dynamics has also been proposed to contribute to evolvability16,17,18, as the conformational energy landscapes of enzymes need to be extensively remodelled upon the emergence of new functions19,20. However, the underlying mechanisms of how protein flexibility affects the emergence of the new and the decline of the original activity are yet to be elucidated18,21. The complete conversion of phosphotriesterase (PTE) to arylesterase (AE) via laboratory evolution22 provides a molecular record to study how dynamics contributes to a 109-fold change in substrate selectivity. During the evolutionary pathway, the new activity improved in a gradually decreasing manner, indicating a slow convergence in function-optimisation, denoted as diminishing returns. The initial 102–103-fold increase in arylesterase activity was accompanied by a moderate decline in the phosphotriesterase activity22, leading to multi-functional intermediates.
Directed and natural evolution of bacterial phosphotriesterases revealed the existence of functionally distinct conformational sub-states12 (Fig. 1). The closed state is optimized for paraoxon hydrolysis, but requires considerable conformational rearrangements for substrate binding (Fig. 1A). In contrast, the open state is poorly organized for the chemical step, but it is optimal for the substrate access (Fig. 1B). Thus, efficient PTE hydrolysis requires an exchange between these two conformational sub-states to compromise between substrate diffusion and conversion23. During the PTE → AE conversion, three PTE loops exhibited substantial changes in dynamics23: L4 (residues 171–175), L5 (residues 202–207) and L7 (residues 258–274) (Fig. 1). L7 fluctuations were gradually frozen out to optimise the new function, while mobilities of L4 and L5 loops increased during the evolution23. Altered dynamics was proposed to shape the landscape of pre-existing catalytic states.
Conformational sub-states in phosphotriesterases with closed (A) and open (B) active sites. (A) The closed configuration (PDB: 2r1n) is optimal for the chemical step, but has a limited substrate access. (B) The open configuration (PDB:3a3w) is easily accessible for the substrate, but less optimised for phosphotriesterase conversion12. The F132–L271 distance (light blue arrow) shrinks from 10 to 4.1 Å from the open to the closed state. The loops are highlighted as L4 (residues 171–175) lime, L5 (residues 202–207) red, and L7 (residues 258–274) blue. The substrate is magenta, the metal ions are displayed by cyan spheres.
Here, we have investigated how the optimisation of functionally distinct conformational sub-states were synchronized in the enzyme dimer, as 6 out of the 26 mutations were located around the dimer interface (within 4.5 Å, Table S1). We observed that the overall change in dynamics, which was estimated by bioinformatics predictions from the sequences of the evolutionary intermediates, correlated to the decline in the original activity. However, we have observed considerable structural deviations between the two enzyme subunits during the PTE → AE trajectory, the magnitude of which has increased with promiscuity. We found that structural divergence was generated by the dynamic coupling between the loops and the dimer interface. Co-evolutionary analysis corroborated the functional link between the loop and interface residues. Taken together, our results highlight the role of protein dynamics via generating asymmetry between the enzyme subunits leading to alternative routes in the emergence of a new function.
Results
Changes in disorder correlate to the new activity
Protein disorder characterizes the preference of a sequence for a well-defined tertiary structure24. Disorder scores can be computed from the primary sequence and provide a coarse description of compactness and flexibility along the protein chain25, in accordance with NMR backbone dynamics26. Thus, disorder scores can used be to estimate the dynamic characteristics from a sequence in the absence of structural data.
We applied this approach to characterize dynamics in all evolutionary intermediates during the PTE → AE conversion, and extended the analysis to those variants whose structures have not been determined. Thus, we used the IUPred method27 to predict the disorder scores for the residues in all evolutionary intermediates22 (R1- R22) and compared them to those in the starting variant (R0) in a pair-wise manner (Methods). First, we computed the root-mean-square deviations of the disorder scores (RMSDID), which characterised the magnitude of the changes in dynamics at a given point of the PTE → AE trajectory (Fig. 2A). We observed that such coarse-grained changes in dynamics correlated to the decrease of the original phosphotriesterase activity (Pearson’s correlations coefficient r = 0.87, significance p = 4.6 × 10–7). Correlation with the emerging arylesterase activity was weaker, but significant (Pearson’s correlations coefficient r = 0.79, significance p = 3.5 × 10–5) (Fig. 2A). These observations indicate that the functional switch PTE → AE requires systematic changes in protein motions.
Changes in dynamics during the PTE → AE conversion. (A) Catalytic efficiencies (kcat/KM) of the original phosphotriesterase (grey) and of the new arylesterase (blue) activity of different evolutionary intermediates as a function of the change in disorder scores (RMSDID) as defined by Eq. (1). The Pearson's r values and their statistical significances are displayed. (B) The impact of mutations on changes in dynamics and structure in different rounds of evolution. The ratio of changes in dynamics [\({RMSD}_{ID }(\mathrm{m})/{RMSD}_{ID }(\mathrm{nm})\)] of mutated and not mutated residues (blue) was defined by Eq. (3), the ratio of structural changes [\({RMSD}_{C\alpha }(\mathrm{m})/{RMSD}_{C\alpha }(\mathrm{nm})\)] was defined by Eq. (4), and was computed separately for the A (light grey) and B (dark grey) subunits. (C) Dynamics changes in coordination shells. The first shell comprises the active site (light blue), the second shell contains residues within 3.5 Å from the active site (dark blue), the third shell residues are within 3.5 Å from the second shell residues (light grey), and the fourth shell residues are the 3.5 Å from the third shell residues (dark grey). Changes in disorder were computed by Eq. (2).
Then, we aimed to probe the contributions of the mutations to the changes in dynamics. Thus, we determined the ratio of RMSDID values, which were computed separately for mutated and not-mutated residues (Methods). As the disordered scores were computed using long flanking windows (Methods), mutations could affect the scores of those residues which were far-lying in sequence. Our results showed that mutations had a larger impact on dynamics than residues which were not changed in evolution (Fig. 2B). In addition, we observed that in the beginning of the PTE → AE conversion mutations affected dynamics more than structure, while towards the end of the evolution this trend was reversed (Fig. 2B). With emerging arylesterase activity (R1–R6), the dynamic contributions of the mutated residues were considerably larger than during specialisation for the new function (R8–R22).
In addition, we observed that overall dynamics increased with promiscuity (R1–R6, Fig. 2C), while exhibited a sharp drop at the generalist state, where original and new activities were comparable. During optimisation of the arylesterase activity (R8–R22) protein dynamics gradually increased, most likely to facilitate structural adjustments for the new function.
Taken together, these results suggest that the emergence of the new function requires significant alterations to protein motions, which enable structural rearrangements for optimising the new activity. This is in accord with the proposed diminishing returns model of evolution22.
Coupling between the loops and the interface
In this section, we investigated how the changes in dynamics were synchronized in the two enzyme subunits during the evolution of the arylesterase function. Thus, we computed the covariance between the disorder scores throughout the PTE → AE conversion using all evolutionary intermediates (Methods) (Figs. 3A, 3B). We observed strong dynamic couplings in the first part of the PTE → AE conversion, when the new function emerged (Fig. 3A). The dynamics of the L4 and L5 loops in particular exhibited a strong positive covariance with increasing promiscuity (R1-R8), while those of the L4 and L7 loops were anti-correlated. This suggests that L4 could serve as a clutch between L5 and L7 loops. The dimer interface exhibited a positive covariance with L4 and L5 dynamics, and a negative covariance with the L7 loop (Fig. 3A). However, we did not observe significant covariance between the loops or with the dimer interface during specialisation for the new function (R9-R22), in accordance with the decrease in dynamical changes in this part of the trajectory (Fig. 3B).
Covariance of dynamics and structure changes during the PTE → AE conversion. (A)–(B) Covariance matrices of changes in disorder scores as compared to the starting R0 variant (defined by Eq. 5), with emergence (A) and specialisation (B) for the new function. (C)–(D) Covariance matrices of deviations in disorder scores and structure from the starting R0 variant (defined by Eq. 6), with emergence (C) and specialisation (D) for the new function. The loops and the dimer interface are indicated on the side panels (L4 green, L5 red, L7 blue, D-dimer interface wheat). Covariance between the loops and the dimer interface are marked by boxes coloured as the corresponding loops. Covariance between the loops are indicated by grey boxes. In the structure-dynamics covariance matrices only the loop structures are displayed for clarity.
We also analysed the covariance between conformation and dynamics in variants whose structures had been determined (R0, R1, R2, R6, R8, R18, R22, Table S3). During the emergence of the new function (R0-R8) we observed a strong, positive covariance between L5 loop structure and L7 dynamics, and a negative covariance between L5 loop structure and L4 dynamics (Fig. 3C). This suggests that changes in dynamics preceded structural rearrangements of the L5 loop. In turn, structural changes of the L5 loop were concomitant with the dynamical changes of the L7 loop. L5 structure was also strongly coupled to the dynamics of the dimeric interface (Fig. 3C). During the optimisation of the new function the structures of both L5 and L4 loops exhibited strong covariance with the interface (Fig. 3D). We observed opposite trends: while changes in dynamics of the interface induced conformational changes in the L5 loop, they decreased structural changes of the L4 loop (Fig. 3D). The L7 loop structure did not exhibit pronounced coupling during the specialisation for the new function.
We performed co-evolutionary analysis on the coupling between the loop and interface residues using the GREMLIN method28 (Methods, Fig. 4, Table S2). We found that residues of the L7 loop and those of the dimer interface exhibit significant co-evolutionary signatures (Fig. 4). In addition, we observed a co-evolution between the L4 and L5 loops, as well as between the N- and C-terminal parts of the dimer interface. These results indicate that dynamical couplings between residues of the loops and the dimer interface, which drive the loop rearrangements, have been evolutionary conserved (Fig. 4).
Co-evolving regions in P. diminuta phosphotriesterase. (A) Co-evolving residues in the L4 (lime), L5 (red), and L7 (blue) loops and the dimer interface (wheat) are shown, as calculated with Gremlin webserver28. Co-evolutionary scores and significances are displayed in Table S2. (B) The structure of the R0 starting variant (PDB:4pcp) with the significant co-evolving residues displayed (black dashed lines). The loops are coloured as L4 lime, L5 red, and L7 blue, and the dimer interface is shown in wheat. The substrate is magenta, the metal ions are displayed by cyan spheres.
Taken together, co-evolutionary results corroborated the functional importance of the dynamical coupling between the loops and the interface residues. Covariance between structure and dynamics indicates that dynamical couplings induce conformational rearrangements for the emergence of a new function.
Functional promiscuity increases conformational divergence between the two subunits
Analysis of the structure-dynamics covariance indicated significant differences between the evolution of the two subunits (Figure S1). In the A subunit, the L5 loop conformation exhibited positive covariance with the dynamics of the L4 and L7 loops and the dimer interface, while negative covariance was observed in the B subunit (Figure S1). These results suggest that dynamic variations, in particular of the dimer interface induced opposite changes in the conformation of the L5 loop in the two subunits. Thus, we have compared the structures of the two subunits in all evolutionary variants with available structures (R0, R1, R2, R6, R8, R18, R22, Table S3). Indeed, we observed that the L5 loop exhibited remarkable deviations between the A and B subunits, especially in the generalist R6 and R8 states (Figure S2).
We further analysed the deviations in L5 conformation in the two subunits and focused on the distances between the loops (Fig. 5). We observed that in subunit B, the separation of the L5 and L7 loops (measured between A/G204 Cα and G273 Cα) decreased substantially during the PTE → AE trajectory, in particular around the generalist R6 and R8 states (Fig. 5, Table S4). In parallel, the L5 loop moved away from L4 in subunit B (measured between G174 Cα and S205 Cα). In subunit A, we observed the opposite trend, the L5–L4 separation decreased, while the distance between the L5 and L7 loops increased slightly with increasing promiscuity (Fig. 5, Table S4). The variations in the L5–L7 distance rationalise why stabilisation of L7 is coupled to destabilisation of L523.
Loop distances during the PTE → AE conversion in the two subunits. (A) Variations in the distance of L5 and L7 loops defined as A/G204 Cα–G273 Cα in the A (light grey) and for the B (dark grey) subunits. (B) Variations in the distance of L4 and L5 loops defined as G174 Cα–S205 Cα in the A (light grey) and for the B (dark grey) subunits. (C) The representative loop positions and their distances in the structure of the B (dark, top) and A subunit (light, bottom) of the generalist R8 variant (PDB:4xay). The loops are coloured as L4 lime, L5 red, and L7 blue; the L5–L7 distance is displayed by a dashed blue line, and the L5–L4 distance by a dashed green line. The substrate is magenta, the metal ions are displayed by cyan spheres.
Taken together, we observed an asymmetry between the dynamical couplings between the loops and the dimer interface, reflecting the opposite movements of the loops in the two subunits.
Alternative evolutionary pathways in the two subunits
We investigated the functional consequences of the different loop positions in the two subunits. First, we compared the two subunits in the generalist R8 intermediate (PDB:4xay) to the structures representing the open (PDB: 3a3w) and closed (PDB:2r1n) states of PTE12 (Figure S3). These structures exhibit some differences in the conformation of the L7 loop, which considerably resize the active size. Although the L5 has a helical conformation in the closed state and is a loop in the open state, its separation from the L4 and L7 loops were rather similar (Figure S3). The position of the L5 loop in the open and closed PTE structures was in between those in the two subunits of the generalist R8 state (Figure S3). These observations indicate that different loop conformations may not be related to the optimisation of substrate binding versus the chemical step.
To obtain further insights into the different loop architectures, we assembled PTE structures from the Protein Data Bank, which represented snapshots of different evolutionary pathways in different organisms. We compared these structures to those of the R6 (PDB: 4xag) and R8 intermediates (PDB:4xay) and analysed the representative distances (L5-L7: A/G204 Cα–G273 Cα; L5-L4: G174 Cα–S205 Cα) between the loops. From the dataset of 160 PTE-related structures we selected 19 structures which had kinetic data on phosphotriesterase or promiscuous activities (Table S5). We observed that enzymes with loop distances similar to the G chain (B subunit) R6 and R8 intermediates, also had considerable arylesterase activity (Table S5), or hydrolysed other substrates (not shown). In those enzymes structures which resemble the loop conformations in the A subunit primarily functioned as phosphotriesterases and were not observed to cleave other substrates. These results suggest that the two subunits of the enzyme dimer have different substrate preferences and catalytic activities in the generalist states. The A subunit is likely specialized on paraoxon hydrolysis, whereas the B subunit is promiscuous, with considerable arylesterase activity.
Taken together, the comparative structural analysis of the subunit conformations to different PTE enzymes indicates that the deviation between the loop architecture has a functional relevance. While the loop conformations in the A subunit are compatible with paraoxon hydrolysis, those in the B subunit have increased preference for arylester hydrolysis. These results implicate that the evolution in the two subunits follows different pathways.
Discussion
The role of conformational heterogeneity in enzymatic catalysis29 as well as in the emergence of new protein functions is being increasingly recognised5. Within this framework, mutations modulate the balance between different conformations, which are compatible with alternative functions16. This sheds light on the role of protein dynamics in protein evolution via modulating conformational ensembles by distant amino acid replacements. Changes in low-frequency structural fluctuations may also facilitate the emergence of new functions7,30, but their interpretation is far from being trivial6. Directed evolution of PTE from Pseudomonas diminuta to arylesterase22 exemplifies how protein evolution engineers protein motions via freezing out fluctuations, which are unproductive for the emerging function.
Our results highlight a novel angle of how catalytic promiscuity stems from conformational heterogeneity. We have found that structural divergence between the two subunits of the Pseudomonas diminuta phosphotriesterase accompanies the evolution of the arylesterase function. Deviation in loop conformations between the two subunits increases with promiscuity. We have shown that dynamic couplings between the loops and the dimer interface, especially during the emergence of the new function, are responsible for the structural divergence (Fig. 3). These results rationalise the role of mutations, which are located at the dimer interface distant from the catalytic centre (Table S1). The functional importance of these dynamic couplings was further corroborated by co-evolutionary data (Fig. 4).
Comparative analysis using a set of PTE enzymes with structures and kinetic data available indicated that the different loop conformations represent functionally distinct states (Table S5). While the loop distances in the A subunit are compatible with phosphotriesterase hydrolysis, in the B subunit (G chain in R6 and R8 intermediates) they are similar to structures with promiscuous activities. Thus, we may conclude that the conversion to arylesterase in the B subunit precedes that of the A subunit. The considerable PTE activity of the A subunit, even at the end of the evolution, was beneficial for the reverse pathway from arylesterase to phosphotriesterase31. Quantifying the difference between the activities of the individual subunits requires state-of-the art QM/MM methods32 because of the metal ions involved in catalysis33 and the heterogeneity of the system34,35, which is being carried out in our laboratory using different simulation methods32.
Taken together, our results indicate that the conversion of PTE to arylesterase is not synchronized in the two subunits of the enzyme dimer. This is analogous to gene duplication; one copy may undergo significant sequence changes, while the other is more restricted to the original function36. Thus, we propose that dimerisation can promote alternative pathways in enzyme evolution via structural divergence due to asymmetric dynamic couplings.
Methods
Disorder predictions
The preference for a well-defined versus a disordered structure was computed using the IUPred program27. Disorder scores correlate to NMR backbone dynamical parameters26, thus they can be used to estimate coarse-grained dynamics at the residue level. We used the sequences of all the intermediates of the PTE → AE conversion22,31 as an input for the IUPred long algorithm to predict the disorder scores. We have also performed predictions using the short version of the IUPred program, the Espritz NMR37 and Dynamine38 methods. The results were found to be consistent with each other.
Characterisation of changes in dynamics
The following quantities were used to assess the impact of evolution on dynamics. The magnitude of the overall changes in dynamics was characterised by the root-mean-square deviation of the ID scores from the starting variant:
where \({RMSD}_{ID}(X)\) is the root-mean square deviation of the disorder scores in the X evolutionary intermediate from the starting (R0) sequence. \({ID}_{i}^{X}\) and \({ID}_{i}^{R0}\) are the ID scores of the ith residue in the X and R0 variant. The RMSD is computed from the scores of all residues, n is the number of residues in the protein.
The change in dynamics in given coordination shells relative to the starting variant was characterised by the sum of the differences between the ID scores of the corresponding residues.
where \({ID}_{i}^{X}\) and \({ID}_{i}^{R0}\) are the ID scores of the ith residue in the X and R0 variant. m is the number of residues in the given (j) coordination shell.
The impact of mutations on dynamics in the X variant was quantified by the ratio of the \({RMSD}_{ID}(X)\) values of the mutated (m) and not mutated (nm) residues:
where \({RMSD}_{ID}(X)\) is the root-mean square deviation of the disorder scores in the X evolutionary intermediate from the starting (R0) sequence. \({ID}_{i}^{X}\) and \({ID}_{i}^{R0}\) are the ID scores of residue i in the X intermediate and the R0 starting variant. Residues were grouped based on whether they were mutated or not in the given round of the PTE → AE conversion. m is the number of mutated, nm is the number of not mutated residues.
Structure analysis. Structures of the evolutionary intermediates were assembled from the Protein Data Bank (Table S3). We noticed a considerable difference between the number and occupancy of the metal ions (Table S3) which are involved in the catalytic step. Structures of the evolutionary intermediates were compared to those of the starting variant (PDB:4pcp). The PTEs with open (PDB: 3a3w) and closed (PDB: 2r1n) active sites (Fig. 1) were derived from earlier studies12. PTE structures with different evolutionary pathways were assembled based on sequence similarity (Table S5).
The impact of mutations on structure in the X variant was quantified by the ratio of the \({RMSD}_{C\alpha }(X)\) values of the mutated (m) and not mutated (nm) residues:
where \({RMSD}_{C\alpha }(X)\) is the root-mean square deviation of the \(C\alpha \) positions in the X evolutionary intermediate from the starting (R0) structure. \({P}_{i,C\alpha }^{X}\) and \({P}_{i,C\alpha }^{R0}\) are the positions of the \(C\alpha \) atom of residue i in the X intermediate and the R0 starting variant. Residues were grouped based on whether they were mutated or not in the given round of the PTE → AE conversion. m is the number of mutated, nm is the number of not mutated residues.
In the analysis of loop distances, the corresponding residue numbers in the PDB structures were identified based on sequence alignment to the wild type PTE sequence.
Covariance analysis
The covariance of changes in dynamics was defined as:
where \({\Delta ID}_{i}\) and \({\Delta ID}_{j}\) are the differences in disorder scores of residues i and j from the starting R0 variant.
The covariance of structure and dynamics changes were defined as:
where \({\Delta ID}_{i}\) is the difference in disorder scores of residue i from the starting R0 variant. \({\Delta C\alpha }_{j}\) is the difference in Cα of residue j from the starting R0 variant.
Covariance matrices were calculated using the R software package (https://www.R-project.org/).
Co-evolution analysis
Co-evolving residue pairs were identified with GREMLIN webserver28 using the wild type PTE22 sequence. The GREMLIN method is based on a multiple sequence alignment (MSA) by HHblits39 with homologue sequences > 90% identity, > 75% coverage and < 25% gaps in MSA. The PTE (A0A060GYS1) co-evolutionary analysis was performed using 819 sequences. Co-evolving amino acids were defined as residue pairs above a scaled score of 1 (Table S2).
References
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Lewandowski, J. R., Halse, M. E., Blackledge, M. & Emsley, L. Protein dynamics. Direct observation of hierarchical protein dynamics. Science 348, 578–581 (2015).
Privett, H. K. et al. Iterative approach to computational enzyme design. Proc. Natl. Acad. Sci. USA 109, 3790–3795 (2012).
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013).
Petrovic, D., Risso, V. A., Kamerlin, S. C. L. & Sanchez-Ruiz, J. M. Conformational dynamics and enzyme evolution. J. R. Soc. Interface 15 (2018).
Kamerlin, S. C. & Warshel, A. At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis?. Proteins 78, 1339–1375 (2010).
Bhabha, G. et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science 332, 234–238 (2011).
Copley, S. D. Shining a light on enzyme promiscuity. Curr. Opin. Struct. Biol. 47, 167–175 (2017).
Khersonsky, O. & Tawfik, D. S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 (2010).
Nam, H. et al. Network context and selection in the evolution to enzyme specificity. Science 337, 1101–1104 (2012).
Aharoni, A. et al. The “evolvability” of promiscuous protein functions. Nat. Genet. 37, 73–76 (2005).
Jackson, C. J. et al. Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase. Proc. Natl. Acad. Sci. USA 106, 21631–21636 (2009).
Allali-Hassani, A. et al. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 5, e97 (2007).
Hong, N. S. et al. The evolution of multiple active site configurations in a designed enzyme. Nat. Commun. 9, 3900 (2018).
Fuxreiter, M. Towards a stochastic paradigm: from fuzzy ensembles to cellular functions. Molecules 23, 3008 (2018).
James, L. C. & Tawfik, D. S. Conformational diversity and protein evolution—a 60-year-old hypothesis revisited. Trends Biochem. Sci. 28, 361–368 (2003).
Tokuriki, N. & Tawfik, D. S. Protein dynamism and evolvability. Science 324, 203–207 (2009).
Campbell, E. C. et al. Laboratory evolution of protein conformational dynamics. Curr. Opin. Struct. Biol. 50, 49–57 (2018).
Zou, T., Risso, V. A., Gavira, J. A., Sanchez-Ruiz, J. M. & Ozkan, S. B. Evolution of conformational dynamics determines the conversion of a promiscuous generalist into a specialist enzyme. Mol. Biol. Evol. 32, 132–143 (2015).
Colin, P. Y. et al. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 6, 10008 (2015).
Pabis, A., Risso, V. A., Sanchez-Ruiz, J. M. & Kamerlin, S. C. Cooperativity and flexibility in enzyme evolution. Curr. Opin. Struct. Biol. 48, 83–92 (2018).
Tokuriki, N. et al. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat. Commun. 3, 1257 (2012).
Campbell, E. et al. The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 12, 944–950 (2016).
van der Lee, R. et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014).
Sormanni, P. et al. Simultaneous quantification of protein order and disorder. Nat. Chem. Biol. 13, 339–342 (2017).
Daughdrill, G. W., Borcherds, W. M. & Wu, H. Disorder predictors also predict backbone dynamics for a family of disordered proteins. PLoS ONE 6, e29207 (2011).
Dosztanyi, Z., Csizmok, V., Tompa, P. & Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347, 827–839 (2005).
Kamisetty, H., Ovchinnikov, S. & Baker, D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc. Natl. Acad. Sci. USA 110, 15674–15679 (2013).
Lu, H. P., Xun, L. & Xie, X. S. Single-molecule enzymatic dynamics. Science 282, 1877–1882 (1998).
Klinman, J. P. & Kohen, A. Evolutionary aspects of enzyme dynamics. J. Biol. Chem. 289, 30205–30212 (2014).
Kaltenbach, M., Jackson, C. J., Campbell, E. C., Hollfelder, F. & Tokuriki, N. Reverse evolution leads to genotypic incompatibility despite functional and active site convergence. Elife 4, e06492 (2015).
Sousa, S. F. et al. Application of quantum mechanics/molecular mechanics methods in the study of enzymatic reaction mechanisms. WIREs Comput. Mol. Sci. 7, e1281 (2017).
Berta, D., Buigues, P. J., Badaoui, M. & Rosta, E. Cations in motion: QM/MM studies of the dynamic and electrostatic roles of H(+) and Mg(2+) ions in enzyme reactions. Curr. Opin. Struct. Biol. 61, 198–206 (2020).
Sanchez-Martinez, M., Marcos, E., Tauler, R., Field, M. & Crehuet, R. Conformational compression and barrier height heterogeneity in the N-acetylglutamate kinase. J. Phys. Chem. B 117, 14261–14272 (2013).
Kots, E. D., Lushchekina, S. V., Varfolomeev, S. D. & Nemukhin, A. V. Role of protein dimeric interface in allosteric inhibition of N-acetyl-aspartate hydrolysis by human aspartoacylase. J. Chem. Inf. Model. 57, 1999–2008 (2017).
Soskine, M. & Tawfik, D. S. Mutational effects and the evolution of new protein functions. Nat Rev Genet 11, 572–582 (2010).
Walsh, I., Martin, A. J., Di Domenico, T. & Tosatto, S. C. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics 28, 503–509 (2012).
Cilia, E., Pancsa, R., Tompa, P., Lenaerts, T. & Vranken, W. F. From protein sequence to dynamics and disorder with DynaMine. Nat. Commun. 4, 2741 (2013).
Remmert, M., Biegert, A., Hauser, A. & Soding, J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2011).
Acknowledgements
M.F. acknowledges the financial support of HAS-11015 and GINOP-2.3.2-15-2016-00044. V.A. is grateful for the one-year fellowship from EFOP-3.6.3-VEKOP-16-2017-00009.
Author information
Authors and Affiliations
Contributions
V.A. performed the disorder calculations, structure and covariance analysis, G.H. assisted the PTE structure analysis and assembly of structural and kinetic data. M.F. designed the study, supervised the work, analysed the results, prepared most of the graphics and wrote the paper.
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ambrus, V., Hoffka, G. & Fuxreiter, M. Asymmetric dynamic coupling promotes alternative evolutionary pathways in an enzyme dimer. Sci Rep 10, 18866 (2020). https://doi.org/10.1038/s41598-020-75772-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75772-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.