Abstract
Several genetic polymorphisms of the haptoglobin gene (HP) or haptoglobin-related gene (HPR) were reported to show a population-specific distribution and to be associated with not only serum haptoglobin (HP) but also cholesterol levels. For such association studies, it is important to know the distribution of polymorphisms or their haplotypes in the populations concerned. However, no comprehensive genetic studies have explored this in Latin Americans, and not every human variation or genotype is available in a database. In this study, we determined the genotypes of common HP (HP1 and HP2), HPdel, rs5471, rs5472, and rs2000999 in several Latin American populations. Haplotypes of rs5472-common HP-rs2000999 polymorphisms were estimated. We did not encounter any HPdel, and the frequencies of rs5471 A, rs5472 A, HP1, and rs2000999 G were higher than their counterpart alleles in studied populations. All of the alleles with higher frequency in the Latin Americans are associated with higher serum HP and lower cholesterol levels. Both A-1-G (probably HP1S) and G-1-G (probably HP1F) haplotypes were higher in Latin American populations than those in other geographic regions. In addition, the genetic influx from populations of other continents into Peruvians seems to be relatively lower than that into other Latin Americans.
Similar content being viewed by others
Introduction
Haptoglobin (HP) is a plasma glycoprotein and is highly expressed in the liver1,2. HP binds hemoglobin (Hb) with a very high affinity to prevent both iron loss and kidney damage due to the oxidative activity of Hb during intravascular hemolysis3. The HP gene (HP) has two codominant alleles of HP1 and HP2 in humans, and as a result, HP has three common phenotypes, HP1-1, HP2-1, and HP2-23.
HP locates on the long arm of chromosome 16 (16q22.3) beside the haptoglobin-related gene (HPR) and consists of five (HP1) or seven (HP2) exons. It has been considered that HP2 is generated by a 1.7-kb intragenic duplication of exons 3 and 4 of HP14. However, a recent study suggested that HP1 likely arose from several recurring deletions of duplicated exons of HP2 by imputation from SNP haplotypes5. In any event, HP1and HP2 are copy number variations (CNVs). HP1 further has two subtypes, variants that migrate faster (HP1F) and slower (HP1S) on starch gel due to two amino acids, positions 52 and 53 (Asp-Lys for 1F, Asn-Glu for 1S), in the duplicated region1. HP2 also is divided into three subtypes: 2FS (containing 1F plus 1S sequences), 2SS (1S plus 1S), 2FF (1F plus 1F). The phenotyping of 1F/S is not easy, and it is also difficult to examine the polymorphic sites responsible for 1F/S directly by fast and simple methods such as real-time PCR due to the complexity of the gene structure1,5. An “indirect” PCR restriction fragment length polymorphism (PCR–RFLP) method has been used to distinguish the SNP associated with the polymorphic sites responsible for 1F/S6,7. We previously analyzed the genetic variation of the HP gene including 1F/S in Ghanaians, Europeans, and Chinese, and found that the A and G alleles of a promoter SNP at position -55 (rs5472) seemed to be almost completely linked with HP1F and HP1S, respectively8.
In addition to common HP polymorphisms, several rare variants of the HP phenotypes have been reported3. One of them is a complete HP deletion allele (HPdel) that has an approximately 28 kb deletion extending from the HP promoter region to intron 4 of HPR9. HPdel homozygotes are at risk for developing anaphylactic transfusion reactions if they produce antibodies (IgE) to HP9,10. This allele has been found only in East and Southeast Asian populations so far8,9,11,12,13,14,15,16,17,18,19.
Because HP is one of the acute phase proteins, its serum level increases in various clinical states such as infectious diseases, malignancy, autoimmune disease, and tissue necrosis. On the other hand, levels decrease during hemolysis, ineffective erythropoiesis, liver disease, and late pregnancy3,20. In addition, gender, age, smoking, plasma Hb levels, serum lipids, and some genetic factors were shown to be associated with circulating HP levels21,22. Serum HP concentrations are reported to be associated with the common HP genotypes and HPdel3,12,23. A SNP located in HPR intron 2 (rs2000999), which was originally identified as one of the genetic determinants of serum total cholesterol by a genome-wide association study (GWAS)24, was thereafter reported to be associated with the serum HP level12,21,22,23,25. A promoter SNP at position − 61 (rs5471), a characteristic SNP of Africans, was identified as a causal polymorphism of HP 2-1 modified phenotypes due to a decreased amount of HP2 polypeptide relative to that of HP1 polypeptide6. Recently we suggested that this SNP is a strong genetic determinant of the HP level in Ghanaians26.
In addition, associations between these polymorphisms as well as rs2000999 and serum cholesterol levels were reported5,23,24,27,28,29,30. Each allele of the polymorphisms that correlated with a higher HP level was associated with lower cholesterol levels despite the population, although no report is available for rs5471 yet.
Recently, human zonulin was identified as a pre-HP2 that enhances intestinal permeability by modulation of intracellular tight junctions31. Thus, the relationship between zonulin and the common HP genotypes has received attention for their potential involvement in the pathogenesis of gastrointestinal diseases and association studies with autoimmune, infective, metabolic, and tumoral diseases such as Celiac disease, obesity, and irritable bowel syndrome32.
Public databases provide human variation and genotype data; however, the distribution of a common HP polymorphism, HPdel, and some SNPs including rs5472 and the polymorphisms responsible for 1F/S (such as rs137853233) are not available in these databases. As seen above, genetic polymorphisms of HP are distributed in a population-specific manner. However, to our knowledge no study so far has explored the comprehensive relationship among the polymorphisms in modern Latin Americans. In this study, to understand genetic polymorphisms in Latin American populations as the basis for an association study, we genotyped for rs5471 and rs5472, which also probably represent 1F/S, common HP alleles, rs2000999, and HPdel.
Results
Development and validation of genotyping for rs5471 and rs5472 by real-time PCR and HRM assays
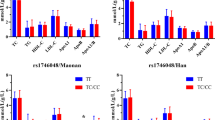
We developed real-time PCR and HRM assays for genotyping the HP promoter polymorphisms rs5471 and rs5472. As mentioned above, rs5471 is a characteristic SNP of Africans, while rs5472 is common in various populations. The frequencies of the C allele of rs5471 and G allele of rs5472 were 12.7% and 41.8%, respectively, in 122 Ghanaian subjects, whose promoter polymorphisms had already been determined by direct sequencing33. Because the rs5471 C allele seems to completely link with the rs5472 A allele, six haplotypes of rs5471 and rs5472 were found in Ghanaians, i.e., AA/AA, AA/CA, AG/AG, AA/AG, AG/CA, and CA/CA. Thus, in order to validate the designed HRM assays, we first examined 122 Ghanaian subjects. As a result, amplicons for rs5471 were divided into four groups, group 1 (all of 94 AA/AA, AA/AG, AG/AG), group 2 (6 of 14 AA/CA), group 3 (8 of 14 AA/CA and all 10 AG/CA), and group 4 (all of 3 CA/CA) (Fig. 1A,B). Amplicons for rs5472 were also divided into four groups, group 1 (all 45 AA/AA, AA/CA, CA/CA), group 2 (26 of 42 AA/AG), group 3 (16 of 42 AA/AG and all of 10 AG/CA), and group 4 (all of 25 AG/AG) (Fig. 1C,D). One individual had a rare base substitution at position of − 68 (rs55663121) as a heterozygote (T/C). The genotype of this individual was A/A at rs5471 and rs5472, and it belonged to group 3 of rs5471and group 1 of rs5472. In any case, we determined six rs5471 and rs5472 haplotypes accurately except one subject (heterozygote of rs55663121) when we comprehensively considered both HRM results for rs5471 and rs5472 (Table 1). We then determined the haplotypes of rs5471 and rs5472 in 416 Latin American individuals (Table 2).
Real-time PCR and HRM analysis of HP promoter polymorphisms. Typical results of amplicon for rs5471 and amplicon for rs5472 on Ghanaians (n = 95) are shown. Normalized and temperature-shifted melting curves of amplicons for rs5471 (A) and rs5472 (C) and normalized and temperature-shifted difference plots for rs5471 (B) and rs5472 (D).
Allele frequency of four polymorphisms and haplotype frequency of three polymorphisms
To know more about the distribution of the HPdel, which has been encountered only in East and Southeast Asia, we screened this allele and common HP polymorphism by TaqMan assay. However, we did not encounter any HPdel allele in the studied populations. The frequency of HP1 is known to be relatively high in Latin Americans3,34. We encountered it at 45.6–100% in this study, but only five samples were available from Mexican Indians (Table 2). We also genotyped rs2000999, and the results are shown in Table 2. The distributions of all polymorphisms were in Hardy–Weinberg equilibrium (HWE) in all populations except rs5472 in the Colombian population (p = 0.00379). However, this value was not significant after Bonferroni correction (adjusted p value = 0.00263, Table 2). In the studied population, five subjects had the rs5471 C allele in a heterozygous state. Whole genome DNA sequencing data of four of the five subjects having the rs5471 C allele were available in a 1,000 genome database (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/), and the results of rs5471 obtained in this study are consistent with those of this database. On the other hand, allele frequencies of HPdel, rs5472, and common HP polymorphisms were not available in this database.
Linkage disequilibrium between three polymorphisms
Because, we found only five of the rs5471 C allele and it seemed to be associated with the A-2-G (rs5472-common-rs2000999) haplotype, we excluded this allele from further haplotype estimations. Thus, we estimated the most likely haplotypes composed of three polymorphisms, rs5472, a common HP polymorphism, and rs2000999 using PHASE software. The deduced haplotypes and their frequencies are shown in Table 3 together with those of Ghanaians, Mongolians, Japanese, and Europeans12. Previous studies suggested that rs5472 G and HP1F and rs5472 A and HP1S were in complete or almost complete linkage disequilibrium in Ghanaian, European, and Chinese populations (also see Table 4 for Ghanaians)8,33. Thus, as in Ghanaians (34.4%), G-1-G probably represented the 1F phenotype and is a characteristic haplotype in Europeans (17.0%). This haplotype was relatively higher (5.0–30.0%) in Latin American populations than in East Asian populations (0.1–2.8%). In addition, the frequency of A-1-G, which probably represented a 1S phenotype, was also higher in Latin American populations (28.1–80%) than in other populations (15.6–27.1%). We then calculated the linkage disequilibrium between each pair of the two polymorphisms and compared the data with those of other populations. As shown in Table 4, the common HP polymorphism and rs2000999 are in strong linkage disequilibrium in all populations (|D′|= 0.924–1, r2 = 0.101–0.375). In addition, rs2000999 is in complete linkage disequilibrium with rs5472 in Latin American populations (|D′|= 1, r2 = 0.250–0.615) and is similar to that of Europeans (|D′|= 1.000, r2 = 0.406), while the linkage disequilibrium between rs5472 and common HP polymorphism is much weaker than those of East Asians and is similar to that of Europeans.
Fixation index (FST) values among five American populations
To quantify population differences, we calculated FST values pairwise among the five populations (Table 5). When following the qualitative guidelines proposed by Wright35, there was little genetic differentiation between populations except Peruvians. On the other hand, there was a moderate differentiation between the Peruvians and Puerto Ricans or Caribbeans.
Discussion
HPdel homozygotes have anhaptoglobinemia, and they are at risk of suffering severe adverse effects of transfusion9,12,36. Thus, in the regions where HPdel is distributed, a genetic test to detect homozygotes before transfusion may be effective to prevent anaphylaxis. However, no study of health problems among homozygotes has been reported, and we are not able to think of any disadvantages caused by having the HPdel allele, except in transfusion, although HP functions as a scavenger of harmful free hemoglobin in intravascular hemolysis, and then HPdel homozygotes might be at a disadvantage.
In our previous study, HPdel was not encountered in Nepalese (Tibetans and Tamang)13. Tibetans and Han Chinese were reported to share relatively recent (7,000–10,000 years ago) Y and mitochondrial DNA haplotypes37,38. If individuals with HPdel had migrated to Tibet 7,000–10,000 years ago, this allele would be distributed in Tibet unless it has a deleterious effect on highlanders or it was neutral in these populations and became extinct due to genetic drift, as described in a previous study18. Considering these findings, the HPdel allele seems to have been generated relatively recently somewhere in China and spread to East and Southeast Asians in a relatively short time period. Our present result that there is no HPdel allele in the Latin American populations also supports this hypothesis.
Our previous studies suggested that the A-1(-G) haplotype represented the 1S phenotype and G-1(-G) represents 1F in African, European, and Chinese populations8,33. This means that we can predict the 1F/S subtype by genotyping rs5472. Because HP2 also contains 1F and/or 1S type sequences, it is difficult to genotype the polymorphisms directly by a conventional method. Thus previous studies, including ours, predicted 1F/S subtypes using indirect methods such as PCR to amplify the relatively large HP1 allele (at least 1.7 kb), followed by restriction enzyme digestion (PCR–RFLP using XbaI or DraI) to recognize a SNP that locates near and links with the polymorphisms responsible for 1F/S7,33. However, it is difficult to examine many samples by PCR–RFLP. A number of association studies between a common HP polymorphism and susceptibility to various diseases and various clinical states have been performed in previous decades3. Recently, high-throughput methods have been reported to impute common HP alleles from SNP data obtained by microarray5,32. As described earlier, several genetic polymorphisms of HP have been reported to be associated with serum HP and cholesterol levels. In addition to recently developed high-throughput methods for imputation of common HP alleles, the haplotype estimation of rs5472 and common HP polymorphisms by real-time PCR assays seemed to be useful for large-scale association studies for HP genotypes particularly including HP1F and HP1S.
We observed relatively higher population differentiation statistics (FST) between Peruvians and other Latin American populations. This may be mainly explained by the lower frequency of G-1-G (5.0% vs. 10.8–30.0%) and G-2-A (9.3% vs. 17.5–20%) haplotypes, and the higher frequency of the A-1-G (53.6% vs. 28.1–39.8%) haplotype. Although the sample size is too small (n = 5), the frequency of G-1-G was relatively lower than that of A-1-G (ratio of G-1-G/A-1-G is 0.25), and there was no HP2 in Mexican Indians. It is speculated that rs5471 A, rs5472 A, HP1, and rs2000999 G alleles and the A-1-G haplotype were prevalent, and HP2 and G-1-G haplotypes (this means HP 1F and mainly migrations from Europe and Africa) were rare in Native Americans. Interestingly, all of the alleles with higher frequency in the Latin Americans are associated with higher serum HP and lower cholesterol levels5,23,24,27,28,29,30. In addition, the genetic influx from populations of other continents into Peruvians seems to be relatively lower than into other Latin Americans, as suggested by previous studies on autosomal, X-, or Y-chromosomal and mitochondrial markers or SNPs or our FUT2 data on the same subjects39,40,41.
The limitations of our study are as follows: (1) the sample sizes are too small to determine the precise allelic frequency of each polymorphism and to conclude that the HPdel is absent in these populations. (2) We could not examine the HP phenotype or serum HP concentration because only DNA samples were available. (3) We did not determine HP1F/S status of the studied samples.
Materials and methods
This study protocol was approved by the Ethical Committee of Kurume University, Japan.
Subjects
Genomic DNA from 122 randomly selected Ghanaians, whose promoter polymorphisms were already determined by direct sequencing analysis, was isolated as described in a previous study33. A total of 416 genomic DNA samples from four 1,000 Genomes project—panels (70 Puerto Ricans in Puerto Rico, MGP00004; 70 Colombians in Medellin, MGP00005; 71 of Mexican ancestry in Los Angeles, MGP00006; 70 Peruvians in Lima, MGP00011), 100 Mexican-Americans in Los Angeles, HD100MEX-2; 10 Human Variation Panel-Mexicans (mixture of seven Mexicans and three Mexican Americans), HD08; 10 Human Variation Panel-Puerto Ricans, HD09; 10 Human Variation Panel-Caribbeans. HD14; and 5 Human Variation Panel-Mexican Indians (Pima Indians from northwest Mexico), HD28 were purchased from the Coriell Institute for Medical Research (Camden, NJ, USA). Because the origin of the Caribbeans is unclear, we treated them as an independent population group. In total, we grouped the populations into six population groups (Table 2).
Genotyping of polymorphisms
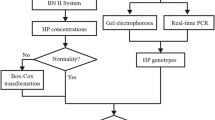
The zygosity of HPdel in addition to that of common HP alleles was determined using a previously described TaqMan assay42. Genotyping of a SNP, rs2000999, was performed as described previously12. Briefly, real-time PCR was carried out in 10 μl of 1 × universal probe master (FastStart, Roche Diagnostics, Tokyo, Japan) containing 0.08 μl of a predesigned TaqMan SNP genotyping assay (Assay ID C_11439045_10, ThermoFisher Scientific, Tokyo, Japan). The temperature profile was 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 45 s. Because previous studies suggested a genetic influx from Africans into modern Latin American populations, we genotyped an African-specific SNP, rs5471, in this study8,39,40,41. Real-time PCR and high-resolution melt (HRM) assays were performed for genotyping two SNPs, rs5471 and rs5472. The primer pairs and amplicons for detection of rs5471 and rs5472 polymorphisms are indicated in Fig. 2. We scanned for amplicon rs5471 of the 63 bp region, and amplicon rs5472 of the 52 bp region. Because rs5471 and rs5472 were located only 6 bp apart, we designed a reverse primer for rs5471 containing T at rs5472 and a forward primer for rs5472 containing A at rs5471 (Fig. 2). PCR amplification and HRM analysis were performed using a real-time PCR platform (LightCycler 480 instrument II, Roche Life Science) and a LightCycler 480 High Resolution Melting Master (Roche Diagnostics) as described previously43. Genotype frequencies were calculated by the counting method and assessed for deviations from HWE by using the exact test. Since the standard exact p value is overly conservative for small minor allele frequencies, we use the mid p value to improve this problem44. Maximum-likelihood haplotype frequencies were estimated using PHASE (version 2.1.1)45.
Estimation of pairwise linkage disequilibrium and FST of genetic differentiation
Pairwise linkage disequilibrium (LD) between polymorphisms and population differentiation statistics (FST) were calculated from the haplotype frequency data of HP by using the DnaSP 6.12.03 software package46.
References
Bowman, B. H. & Kurosky, A. Haptoglobin: The evolutionary product of duplication, unequal crossing over, and point mutation. Adv. Hum. Genet.12(189–261), 453–184 (1982).
Wicher, K. B. & Fries, E. Haptoglobin, a hemoglobin-binding plasma protein, is present in bony fish and mammals but not in frog and chicken. Proc. Natl. Acad. Sci. USA103, 4168–4173. https://doi.org/10.1073/pnas.0508723103 (2006).
Carter, K. & Worwood, M. Haptoglobin: A review of the major allele frequencies worldwide and their association with diseases. Int. J. Lab. Hematol.29, 92–110. https://doi.org/10.1111/j.1751-553X.2007.00898.x (2007).
Maeda, N., Yang, F., Barnett, D. R., Bowman, B. H. & Smithies, O. Duplication within the haptoglobin Hp2 gene. Nature309, 131–135 (1984).
Boettger, L. M. et al. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat. Genet.48, 359–366. https://doi.org/10.1038/ng.3510 (2016).
Maeda, N. DNA polymorphisms in the controlling region of the human haptoglobin genes: A molecular explanation for the haptoglobin 2–1 modified phenotype. Am. J. Hum. Genet.49, 158–166 (1991).
Koch, W. et al. Haptoglobin gene subtyping by restriction enzyme analysis. Clin. Chem.49, 1937–1940. https://doi.org/10.1373/clinchem.2003.022442 (2003).
Teye, K. et al. Haptoglobin gene promoter polymorphism and haplotypes are unique in different populations. Hum. Biol.78, 121–126 (2006).
Koda, Y. et al. Simple PCR detection of haptoglobin gene deletion in anhaptoglobinemic patients with antihaptoglobin antibody that causes anaphylactic transfusion reactions. Blood95, 1138–1143 (2000).
Shimada, E. et al. Anaphylactic transfusion reactions in haptoglobin-deficient patients with IgE and IgG haptoglobin antibodies. Transfusion42, 766–773. https://doi.org/10.1046/j.1537-2995.2002.00117.x (2002).
Nakamura, H., Soejima, M., Munkhtulga, L., Iwamoto, S. & Koda, Y. Haptoglobin polymorphism in Mongolian population: Comparison of the two genotyping methods. Clin. Chim. Acta408, 110–113. https://doi.org/10.1016/j.cca.2009.08.001 (2009).
Soejima, M. et al. Genetic factors associated with serum haptoglobin level in a Japanese population. Clin. Chim. Acta433C, 54–57. https://doi.org/10.1016/j.cca.2014.02.029 (2014).
Soejima, M., Koda, Y., Fujihara, J. & Takeshita, H. The distribution of haptoglobin-gene deletion (Hp del) is restricted to East Asians. Transfusion47, 1948–1950. https://doi.org/10.1111/j.1537-2995.2007.01467.x (2007).
Park, K. U., Song, J. & Kim, J. Q. Haptoglobin genotypic distribution (including Hp0 allele) and associated serum haptoglobin concentrations in Koreans. J. Clin. Pathol.57, 1094–1095. https://doi.org/10.1136/jcp.2004.017582 (2004).
Su, Y. C., Chen, Y. C., Li, S. C., Lee, C. C. & Tung, Y. T. Detection of Hpdel in healthy individuals and cancer patients in Taiwan. Clin. Chem. Lab. Med.47, 745–749. https://doi.org/10.1515/CCLM.2009.156 (2009).
Soejima, M. & Koda, Y. Rapid real-time PCR detection of HPdel directly from diluted blood samples. Clin. Chem.54, 1095–1096. https://doi.org/10.1373/clinchem.2008.103747 (2008).
Shimada, E. et al. Detection of Hpdel among Thais, a deleted allele of the haptoglobin gene that causes congenital haptoglobin deficiency. Transfusion47, 2315–2321. https://doi.org/10.1111/j.1537-2995.2007.01473.x (2007).
Soejima, M. et al. Haptoglobin genotyping of Vietnamese: Global distribution of HP del, complete deletion allele of the HP gene. Leg. Med. (Tokyo)17, 14–16. https://doi.org/10.1016/j.legalmed.2014.08.004 (2015).
Cox, S. E. et al. Haplotype association between haptoglobin (Hp2) and Hp promoter SNP (A-61C) may explain previous controversy of haptoglobin and malaria protection. PLoS One2, e362. https://doi.org/10.1371/journal.pone.0000362 (2007).
Gabay, C. & Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med.340, 448–454. https://doi.org/10.1056/NEJM199902113400607 (1999).
Froguel, P. et al. A genome-wide association study identifies rs2000999 as a strong genetic determinant of circulating haptoglobin levels. PLoS One7, e32327. https://doi.org/10.1371/journal.pone.0032327 (2012).
Soejima, M., Munkhtulga, L., Furukawa, K., Iwamoto, S. & Koda, Y. Serum haptoglobin correlates positively with cholesterol and triglyceride concentrations in an obese Mongolian population. Clin. Chim. Acta505, 176–182. https://doi.org/10.1016/j.cca.2020.03.003 (2020).
Wang, S. et al. Mendelian randomization analysis to assess a causal effect of haptoglobin on macroangiopathy in Chinese type 2 diabetes patients. Cardiovasc. Diabetol.17, 14. https://doi.org/10.1186/s12933-018-0662-7 (2018).
Igl, W. et al. Modeling of environmental effects in genome-wide association studies identifies SLC2A2 and HP as novel loci influencing serum cholesterol levels. PLoS Genet.6, e1000798. https://doi.org/10.1371/journal.pgen.1000798 (2010).
Shahabi, P., Siest, G., Herbeth, B., Ndiaye, N. C. & Visvikis-Siest, S. Clinical necessity of partitioning of human plasma haptoglobin reference intervals by recently-discovered rs2000999. Clin. Chim. Acta413, 1618–1624. https://doi.org/10.1016/j.cca.2012.04.033 (2012).
Soejima, M., Teye, K. & Koda, Y. The haptoglobin promoter polymorphism rs5471 is the most definitive genetic determinant of serum haptoglobin level in a Ghanaian population. Clin. Chim. Acta483, 303–307. https://doi.org/10.1016/j.cca.2018.05.029 (2018).
Zheng, N. S. et al. A common deletion in the haptoglobin gene associated with blood cholesterol levels among Chinese women. J. Hum. Genet.62, 911–914. https://doi.org/10.1038/jhg.2017.66 (2017).
Saha, N., Liu, Y., Tay, J. S., Basair, J. & Ho, C. H. Association of haptoglobin types with serum lipids and apolipoproteins in a Chinese population. Clin. Genet.42, 57–61 (1992).
Braeckman, L., De Bacquer, D., Delanghe, J., Claeys, L. & De Backer, G. Associations between haptoglobin polymorphism, lipids, lipoproteins and inflammatory variables. Atherosclerosis143, 383–388 (1999).
Guthrie, P. A. et al. Complexity of a complex trait locus: HP, HPR, haemoglobin and cholesterol. Gene499, 8–13. https://doi.org/10.1016/j.gene.2012.03.034 (2012).
Fasano, A. Zonulin, regulation of tight junctions, and autoimmune diseases. Ann. N. Y. Acad. Sci.1258, 25–33. https://doi.org/10.1111/j.1749-6632.2012.06538.x (2012).
Cupaioli, F. A. et al. Assessment of haptoglobin alleles in autism spectrum disorders. Sci. Rep.10, 7758. https://doi.org/10.1038/s41598-020-64679-w (2020).
Teye, K. et al. A-61C and C-101G Hp gene promoter polymorphisms are, respectively, associated with ahaptoglobinaemia and hypohaptoglobinaemia in Ghana. Clin. Genet.64, 439–443 (2003).
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. The History and Geography of Human Genes. 204 Maps (Princeton University Press, Princeton, 1994).
Wright, S. Evolution and the Genetics of Populations, Vol. 4 (University of Chicago Press, Chicago, 1978).
Koda, Y., Soejima, M., Yoshioka, N. & Kimura, H. The haptoglobin-gene deletion responsible for anhaptoglobinemia. Am. J. Hum. Genet.62, 245–252. https://doi.org/10.1086/301701 (1998).
Gayden, T. et al. The Himalayas: Barrier and conduit for gene flow. Am. J. Phys. Anthropol.151, 169–182. https://doi.org/10.1002/ajpa.22240 (2013).
Qi, X. et al. Genetic evidence of Paleolithic colonization and Neolithic expansion of modern humans on the Tibetan Plateau. Mol. Biol. Evol.30, 1761–1778. https://doi.org/10.1093/molbev/mst093 (2013).
Soejima, M. & Koda, Y. FUT2 polymorphism in Latin American populations. Clin. Chim. Acta505, 1–5. https://doi.org/10.1016/j.cca.2020.02.011 (2020).
Adhikari, K., Mendoza-Revilla, J., Chacón-Duque, J. C., Fuentes-Guajardo, M. & Ruiz-Linares, A. Admixture in Latin America. Curr. Opin. Genet. Dev.41, 106–114. https://doi.org/10.1016/j.gde.2016.09.003 (2016).
Chacón-Duque, J. C. et al. Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nat. Commun.9, 5388. https://doi.org/10.1038/s41467-018-07748-z (2018).
Soejima, M. & Koda, Y. TaqMan-based real-time PCR for genotyping common polymorphisms of haptoglobin (HP1 and HP2). Clin. Chem.54, 1908–1913. https://doi.org/10.1373/clinchem.2008.113126 (2008).
Soejima, M. et al. Genetic variation of FUT2 in a Vietnamese population: Identification of two novel Se enzyme-inactivating mutations. Transfusion52, 1268–1275. https://doi.org/10.1111/j.1537-2995.2011.03485.x (2012).
Graffelman, J. & Moreno, V. The mid p-value in exact tests for Hardy-Weinberg equilibrium. Stat. Appl. Genet. Mol. Biol.12, 433–448. https://doi.org/10.1515/sagmb-2012-0039 (2013).
Stephens, M., Smith, N. J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet.68, 978–989. https://doi.org/10.1086/319501 (2001).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol.34, 3299–3302. https://doi.org/10.1093/molbev/msx248 (2017).
Acknowledgements
We thank Ms. Katherine Ono for the English editing of this manuscript. This work was supported in part by JSPS KAKENHI Grant number JP17K09283.
Author information
Authors and Affiliations
Contributions
M.S. performed experiments and contributed to the data analysis and drafting of the manuscript. Y.K. planned the experiment, contributed to the data analysis and interpretation, and drafted the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Soejima, M., Koda, Y. Haptoglobin polymorphisms in Latin American populations. Sci Rep 10, 13780 (2020). https://doi.org/10.1038/s41598-020-70755-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70755-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.