Abstract
A 360-area surface-based cortical parcellation is extended to study mild cognitive impairment (MCI) and Alzheimer’s disease (AD) from healthy control (HC) using the joint human connectome project multi-modal parcellation (JHCPMMP) proposed by us. We propose a novel classification method named as JMMP-LRR to accurately identify different stages toward AD by integrating the JHCPMMP with the logistic regression-recursive feature elimination (LR-RFE). In three-group classification, the average accuracy is 89.0% for HC, MCI, and AD compared to previous studies using other cortical separation with the best classification accuracy of 81.5%. By counting the number of brain regions whose feature is in the feature subset selected with JMMP-LRR, we find that five brain areas often appear in the selected features. The five core brain areas are Fusiform Face Complex (L-FFC), Area 10d (L-10d), Orbital Frontal Complex (R-OFC), Perirhinal Ectorhinal (L-PeEc) and Area TG dorsal (L-TGd, R-TGd). The features corresponding to the five core brain areas are used to form a new feature subset for three classifications with the average accuracy of 80.0%. Results demonstrate the importance of the five core brain regions in identifying different stages toward AD. Experiment results show that the proposed method has better accuracy for the classification of HC, MCI, AD, and it also proves that the division of brain regions using JHCPMMP is more scientific and effective than other methods.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the most common type of neurodegenerative disorder characterized by progressive impairment of memory and other cognitive functions in elderly people worldwide, and results in elderly people to death eventually. Pre-clinical stage of Alzheimer’s disease, also known as mild cognitive impairment (MCI), is a transitional state between normal aging and AD, often an early warning signal of AD. The correct recognition of MCI and AD plays an important role in the prevention, early detection and intervention of AD, and lays a foundation for the exploration of effective treatment methods for AD in the future.
The Human Connectome Program (HCP) proposed a multi-modal parcellation (MMP)1 of the human cerebral cortex with 180 areas per hemisphere. The HCPMMP is based on surface-registered multi-modal MR acquisition and objective semi-autonomic nerve anatomy, and the criteria are sharp changes in cortical architecture, function and connectivity. A range of studies2,3,4 have demonstrated that the widespread application of HCPMMP can help to understand the healthy brain and dementia, such as AD, schizophrenia (SCZ), Parkinson’s disease (PD). Some studies have shown that the brain connectivity for neurodegenerative diseases has changed significantly, and the topological structure of the brain network has been disrupted4,5,6,7,8. Network measurement of different regions of the human brain is considered to be an effective feature for recognition of cognitive impairment patients9. The HCPMMP sample was derived from 210 healthy adults but there are few studies on the cerebral cortex of AD patients. JHCPMMP introduced HCPMMP into the cerebral cortex of AD patients and applied it to the classification of HC, MCI, and AD10. The main goal of our study is to demonstrate an automated and accurate method for identification of AD, MCI and HC.
Method
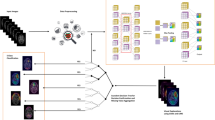
A novel classification approach is proposed to accurately identify different stages toward AD by integrating the JHCPMMP with the logistic regression-recursive feature elimination (LR-RFE), which is named as JMMP-LRR. This method is applied to complete the entire experiment. Firstly, the sparse network is obtained by using JHCPMMP10. The process of this step is to process the fMRI data, project it to CIFTI Space, and obtain the sparse network through MMP. Secondly, we calculate the 9 attributes of brain networks, including strength, betweenness centrality, local efficiency etc, and obtain 3,240 candidate features of each subject. Subsequently, we apply LR-RFE to select the 30 features of each subject. Finally, the classifier of OVR-SVM is applied to classify the extracted features of HC, MCI and AD for classification. The process of the three-class classification in this paper is shown in Fig. 1.
Data preprocessing
The brain is parcellated with 180 areas per hemisphere by using HCPMMP atlas, which delineates the cortical architecture, function, and connectivity. The sparse network is obtained with the help of JHCPMMP10. The process of this step is to process the fMRI data, project it to CIFTI Space, and obtain the sparse network through MMP. MMP can show dramatic changes in cortical thickness, myelin atlas, task fMRI, and resting fMRI for each brain region. The correlation can be calculated for 360 areas. The sparse network is generated by searching the proportion of the strongest weights (PSW). The purpose of this step is to reduce noise and weakly correlated connections.
Network features in each node of the connectivity network are calculated as the candidate features. The feature vector of each sample contained strength (S), betweenness centrality (BC), clustering coefficient (CC), local efficiency (LE), eigenvector centrality (EC), k-coreness centrality (KC), page rank centrality (PC), Subgraph centrality (SC) and flow coefficient (FC). The software calculating graph theoretical measures can be the Brain Connectivity Toolbox (BCT, available at: https://sites.google.com/site/bctnet/).
For single local network measure, a vector of 360 × 1 is formed in which each vector represents an eigenvalue from the corresponding functional area in brain cortex. By calculating the attributes of the brain network, a feature matrix of 360 × 9 is formed, and each feature is stored in a column. The advantage of scaling each column of the eigenvalue matrix is that the range of the eigenvalue is not too large, which leads to the dominance of the more valuable features in classification. Each feature is normalized to the range−1 1. Therefore, the 360 × 9 = 3,240 candidate features are generated for classification of HC, MCI and AD.
Feature selection
The network-based measure generates the 3,240 candidate feature values for classification, which greatly affects the calculation cost and classification accuracy. Then, the 3,240 candidate features was a feature vector for each subject. Noisy and irrelevant features often lead to over-fitting problem. Generally, feature selection should be implemented before classification by extracting a subset of feature from the original 3,240 candidate features, which could reduce training time, test time and improve classification performance.
There are two main methods for feature selection, including filter, wrapper. The characteristic of filter feature selection is to select features from data first, and then train the learner, the process of feature selection is independent of subsequent learners. The wrapper method uses an inductive algorithm directly to evaluate the feature subset, which is generally better than filter method in terms of prediction accuracy, but usually more computationally intensive.
Recursive feature elimination (RFE) is a common method in wrapper feature selection. From the final performance of the learner, the wrapped feature selection is better than the filtered feature selection. The RFE method continuously eliminates the features with low contribution scores on the basis of the iterative method, and then ranks each feature in each cycle to delete the n features with the lowest score.

Pseudo-code for the Recursive Feature Elimination (RFE) algorithm.
LR-RFE algorithm is applied to extract important features from the 3,240 features. The main idea of LR-RFE algorithm is to repeatedly eliminate features with low contribution scores based on the iterative method, and rank each feature in each cycle using LR algorithm to delete the 10 features with the lowest score. The process is repeated for the remaining features until all features are traversed. From the 3,240 features, 30 optimal feature subsets are selected by using LR-RFE. The LR-RFE algorithm is implemented for finding optimal feature subset in Python using the Sklearn package.

LR-RFE algorithm steps.
SVM classifier
One-vs-the-rest support vector machine (OVR-SVM) is applied to achieve high classification accuracy after the dimension of the features has been reduced by LR-REF. SVM is a binary classification model to find a hyperplane to segment the samples. Dealing with multi-class classification problems requires the construction of a suitable multi-class classifier. This paper adopts OvR multi-class strategy, also known as one-vs-all.
OvR is the most commonly used strategy for multi-class classification. One class at a time is taken as a positive example, and the other classes are taken as a negative example to train N classifiers. If only one classifier is predicted as a positive class, the corresponding class label is used as the final classification result. The OvR-SVM is a multivariate statistical method that can be used for classification. In this paper, we use OVR-SVM as the classifier.
The mathematical principle of OVR-SVM is as follows: When you want to distinguish K classes, the problem can be expressed as the mathematical problem described in Eqs. 1–311.
where the training data xi are mapped to a higher dimensional space by the function ϕ and C is the penalty parameter.
When Eq. 7 is solved, there are decision k functions:
We find the largest value of the decision function in the class
Through feature selection, the number of data samples in the experiment is 72 and the number of features is 30, which is consistent with the characteristics of small sample and high dimension. It also indicates that OVR-SVM is very suitable for the three-class classification.
In this process, we use three two-class classifiers, the first two-class classifier is HC as the case, MCI and AD as the counterexample, the second two-class classifier is MCI as the case, HC and AD as the counterexample, the third two-class classifier is AD as the case, HC and AD as the counterexample. Figure 2 shows the process to distinguish AD from HC, MCI, and AD.
We also carry out two-two classifications for HC, MCI, and AD by usingthe SVM algorithm.
Classification and performance metrics
In a pair of training and testing groups, high classification rates may be contingent, so in order to evaluate the prediction performance of the model and reduce over-fitting, we cross-validate the data by 5 folds. The principle of K-fold cross-validation is to divide the whole data into k parts of equal size. Using the model of other k-1 subsets to train classifiers, one of the K parts is tested12. In this experiment, the evaluation model uses the following evaluation indicators: Accuracy, Precision, Recall, F1-score. Table 1 lists the confusion matrix of three classification. Each performance is defined in Eqs. 4–7.
Results
The brain MR imaging data of 72 subjects (mean age:76.3 ± 7.7 years, range: 55.8–95.9 years, meal/female: 40/32) used in this paper are obtained from the Alzheimer’s disease Neuroimaging Initiative (ADNI database (adni.loni.usc.edu), including T1 and T2 structure data, resting state fMRI with eyes open, field map. In the present study, 24 subjects per groups in three classes of HC, MCI and AD were analyzed in this study. Table 2 lists the demographics of all this subjects.
In this paper, the final feature vectors, which are obtained after dimension reduction using LR_REF, are classified by SVM. A total of 2,160 feature vectors (72 subjects × 30 features) are used for classification. The state recognition of HC, MCI, AD is performed with the three two-class SVM classifiers. We use the SVM classifier which is implemented by and choose Linear as kernel.The parameters of SVM are determined by 5-fold cross-validation method. The classification results are summarized in Table 3. As can be seen form the Table 3, the OVR-SVM classifier achieved the accuracy of 89% for classification of three groups of HC, MCI, and AD. Moreover, we further applied two typical methods, namely, logistic regression (LR) and K-nearest neighbor (KNN) in Alzheimer’s disease recognition to the same imaging data for a comprehensive comparison. The classification results are summarized in Table 3, which shows that the proposed method achieves better performance than other two methods. The AD vs. MCI vs. HC classification performance metrics are showed in Fig. 3.
To estimate the generalization ability of our proposed method, experiments are also performed on three binary classification tasks (HC vs. AD, MCI vs. AD, and HC vs. MCI). The classification accuracies of two classes are 98.0% for AD vs. HC, 92.0% for MCI vs. AD, and 95.5% for HC vs. MCI. Similarly, we further applied logistic regression (LR) and K-nearest neighbor (KNN) to the same imaging data for two-class classification as a comparison. The classification results are summarized in Table 4, which shows that the proposed method achieves better performance than other two methods.
The brain regions corresponding to the 30 features involved in classification. With HCPMMP’s rules for dividing brain regions, the number of the brain region in the right brain is 1–180, and the number of the brain region in the left brain is 181–360. Because the brain is symmetrical, the brain region of the left brain can also be found in the right brain.

The step of calculating the brain region.
In the three-class classification and two-classification of Alzheimer’s disease, we used the 30 features corresponding to the 24 cortical areas in Table 5.
As shown in Table 5, we further analyzed the information of 30 features and then found the five key cortical areas, and each and each cortex area corresponded to two or more features, namely Fusiform Face Complex (L-FFC), Area 10d (L-10d), Orbital Frontal Complex (R-OFC), Perirhinal Ectorhinal (L-PeEc) and Area TG dorsal (L-TGd,R-TGd). The corresponding characteristics of specific key areas are shown in Table 6. Their specific distribution in the brain is shown in Fig. 4.
In order to further analyze the five core Cortical areas, the 11 features corresponding to the five Cortical areas of FFC, 10d, OFC, PeEc and TGd are selected from 30 features corresponding to the 24 Cortical areas, which are used to classify HC, MCI, and AD. Subsequently, we use the 5-fold cross-validation of SVM and LR to classify these separately. From Table 7, the accuracies of the classification in SVM and LR with 11 features are 80% and 78%, respectively.
In addition, the accuracies of the classification in SVM and LR with 30 features of 24 cortical areas are 89% and 88%, respectively. Furthermore, in order to analyze the role of the features of five cortical areas in classification, we randomly select the corresponding features of the five cortical areas in the remaining 19 cortical areas to calculate the accuracy of classification. The training and test are repeated 10 times to get the average accuracies for SVM and LR. The classification results of Accuracy_3 were given in Table 7.
From Table 7, the classification accuracies of Set 2 are closer to that of Set 1, but the classification accuracies of Set 3 are much lower than that of Set 1. Obviously, when the features are taken from five Cortical areas of FFC, 10d, OFC, PeEc and TGd, the classification accuracy is high than random five cortical areas. Therefore, we observe that the five cortical areas have a great impact on the results of the three-class classification.
Discussion
Most previous studies focused on the two-class classification between HC, MCI, and AD, and they have achieved great accuracy. With the imaging data of ADNI database, some studies also reported recognition results of three-class classification between HC, MCI, and AD. As shown in Table 8, our method obtained higher accuracy than previous studies using old brain parcellation methods. It shows that our parcellation scheme benefits the classification of HC, MCI, and AD. It also proves that the division of brain regions of JHCPMMP is more scientific and effective than other methods.
As shown in Table 7, when the features are taken from five cortical areas of FFC, 10d, OFC, PeEc and TGd, the classification accuracy is high than that using five random cortical areas. Therefore, the five cortical areas have a great impact on the results of the three-class classification. And this finding has been confirmed in previous clinical papers.
Zebrowitz13 observed lower activation, specificity, and resting blood flow for older adults than younger adults in the fusiform face area (FFA) but not in other regions of interest, and then the facial selection mechanism of the elderly was uncoordinated. Bludau et al.14 found that Fp1 and Fp2 have different contributions to functional networks. Fp1 was involved in cognition, working memory and perception, whereas Fp2 was part of brain networks underlying affective processing and social cognition. Grabenhorst et al.15 pointed out that OFC can affect people’s function of feeling happiness, pain, and reward and punishment. Ding et al.16 found that human TPC actually includes anterior parts of areas 35, 36, and TPC seems to be involved in social and emotional processing to a large extent, including facial processing, recognition and semantic memory. Olson et al.17 studied that TGd may combine complex and highly processed perceptual input with visceral emotional response. Thus, there five areas all have been confirmed to be involved in human facial processing, emotional perception and memory function. Therefore, our results were in line with those reported in previous studies, showing significant importance to further explore the treatment strategies of Alzheimer’s disease, and carry out early intervention to delay the deterioration of the disease.
Conclusion
We propose a method JMMP-LRR which combines LR-RFE and JHCPMMP for three classifications of AD patients. fMRI data is processed by JHCPMMP to obtain small samples, ultra-high-dimensional data, these data directly involved in classification will cause too long running time and low classification accuracy, JMMP-LRR can solve the problem very well. The features obtained by using LR-RFE as feature extraction were more recognizable for the three classifications of AD patients, and could achieve high classification accuracy. By analyzing the features obtained by LR-RFE, we find 5 brain regions were sensitive to AD patient identification: L-FFC, L-10d, R-OFC, L-PeEc, (L-TGd, R-TG). Only use the functional features of these 5 brain regions, we could achieve high accuracy. The accuracies of the two experiments using the JMMP-LRR method were higher than the current method. It also proves that JHCPMMP is better than other brain partitioning methods in identifying patients with AD.
Data availability
Data collection and sharing for this project is funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health, USA).
References
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536.7615, 171 (2016).
Glasser, M. F. et al. The human connectome project’s neuroimaging approach. Nature neuroscience 19(9), 1175 (2016).
Liu, J. et al. Complex brain network analysis and its applications to brain disorders: a survey. Complexity 2017 (2017).
van den Heuvel, M. P. & Yeo, B. T. A spotlight on bridging microscale and macroscale human brain architecture. Neuron 93(6), 1248–1251 (2017).
Khazaee, A., Ebrahimzadeh, A. & Babajani-Feremi, A. Application of advanced machine learning methods on resting-state fMRI network for identification of mild cognitive impairment and Alzheimer’s disease. Brain imaging and behavior 10(3), 799–817 (2016).
Supekar, K., Menon, V., Rubin, D., Musen, M. & Greicius, M. D. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS computational biology 4(6), e1000100 (2008).
Wang, K. et al. Altered functional connectivity in early Alzheimer’s disease: A resting‐state fMRI study. Human brain mapping 28(10), 967–978 (2007).
Liu, Y. et al. Regional homogeneity, functional connectivity and imaging markers of Alzheimer’s disease: a review of resting-state fMRI studies. Neuropsychologia 46(6), 1648–1656 (2008).
Mammone, N. et al. Permutation disalignment index as an indirect, EEG-based, measure of brain connectivity in MCI and AD patients. International journal of neural systems 27(5), 1750020 (2017).
Sheng, J. et al. A novel joint HCPMMP method for automatically classifying Alzheimer’s and different stage MCI patients. Behavioural Brain Research 365, 210–221 (2019).
Suk, H. I. & Shen, D. Deep learning-based feature representation for AD/MCI classification. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 583–590 (2013).
Andrés, O. et al. Disease Neuroimaging Initiative. Exploratory graphical models of functional and structural connectivity patterns for Alzheimer’s disease diagnosis. Frontiers in computational neuroscience 9, 132 (2015).
Zebrowitz, L., Ward, N., Boshyan, J., Gutchess, A. & Hadjikhani, N. Dedifferentiated face processing in older adults is linked to lower resting state metabolic activity in fusiform face area. Brain research 1644, 22–31 (2016).
Bludau, S., Eickhoff, S. B. & Mohlberg, H. Cytoarchitecture, probability maps and functions of the human frontal pole. Neuroimage 93, 260–275 (2014).
Fabian, G. et al. A common neural scale for the subjective pleasantness of different primary rewards. Neuroimage 51(3), 1265–1274 (2010).
Ding, S. L., Van Hoesen, G. W., Cassell, M. D. & Poremba, A. Parcellation of human temporal polar cortex: a combined analysis of multiple cytoarchitectonic, chemoarchitectonic, and pathological markers. Journal of Comparative Neurology 514.6, 595–623 (2009).
Olson, I. R., Plotzker, A. & Ezzyat, Y. The enigmatic temporal pole: a review of findings on social and emotional processing. Brain 130.7, 1718–1731 (2007).
Li, F. et al. A robust deep model for improved classification of AD/MCI patients. IEEE journal of biomedical and health informatics 19(5), 1610–1616 (2015).
Khedher, L., Ramírez, J., Górriz, J. M., Brahim, A. & Illán, I. A. Independent component analysis-based classification of Alzheimer’s disease from segmented MRI data[C]//International Work-Conference on the Interplay between Natural and Artificial Computation. Springer, Cham 78–87 (2015).
Quintana, M. et al. Using artificial neural networks in clinical neuropsychology: High performance in mild cognitive impairment and Alzheimer’s disease. Journal of Clinical and Experimental Neuropsychology 34(2), 195–208 (2012).
Zhang, Y. et al. Detection of Alzheimer’s disease and mild cognitive impairment based on structural volumetric MR images using 3D-DWT and WTA-KSVM trained by PSOTVAC. Biomedical Signal Processing and Control 21, 58–73 (2015).
Tong, T. et al. Multi-modal classification of Alzheimer’s disease using nonlinear graph fusion. Pattern recognition 63, 171–181 (2017).
Lama, R. K., Gwak, J., Park, J.-S. & Lee, S.-W. Diagnosis of Alzheimer’s disease based on structural MRI images using a regularized extreme learning machine and PCA features. Journal of healthcare engineering 2017 (2017).
Son, S. J., Kim, J. & Park, H. Structural and functional connectional fingerprints in mild cognitive impairment and Alzheimer’s disease patients. PloS one 12(3), e0173426 (2017).
Hsu, C. W. & Lin, C. J. A comparison of methods for multiclass support vector machines. IEEE transactions on Neural Networks 13(2), 415–425 (2002).
Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Ijcai 14(2), 1137–1145 (1995).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 61871168).
Author information
Authors and Affiliations
Contributions
J. Sheng designed the project and supervised the overall research; M. Shao performed the experiments and analyzed data; Q. Zhang co-designed the research; R. Zhou, L. Wang, and Y. Xin participated in data analysis. J. Sheng and M. Shao wrote the manuscript. All authors provided feedback and agreed on the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sheng, J., Shao, M., Zhang, Q. et al. Alzheimer’s disease, mild cognitive impairment, and normal aging distinguished by multi-modal parcellation and machine learning. Sci Rep 10, 5475 (2020). https://doi.org/10.1038/s41598-020-62378-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62378-0
This article is cited by
-
Dissecting Psychiatric Heterogeneity and Comorbidity with Core Region-Based Machine Learning
Neuroscience Bulletin (2023)
-
Predictive classification of Alzheimer’s disease using brain imaging and genetic data
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.