Abstract
Oenocarpus distichus presents economic, ecological, and dietary potential for pulp market processed in natura. Germplasm conservation and genetic improvement depend on genetic divergence studies. The objective was to quantify genetic divergence in a native population of O. distichus genotypes based on fruit biometrics. The fruit length and width, fruit mass, pulp and seed, pulp and almond thickness, and pulp yield per fruit were evaluated. All fruit biometric characteristics of O. distichus palms show genetic variability. Genetic variations among genotypes are essential for predicting heredity and heterosis, which are essential for improving O. distichus production. Pulp yield and seed mass were negatively correlated. Almond thickness and pulp, seed mass and transverse diameter were positively correlated. Genetic distances between pair of genotypes ranged from 0.07 to 48.10 with three genetically distinct groups. The seed mass, almond thickness and transverse diameter contributed to genetic divergence. Heritability estimates the genetic control that can be obtained from O. distichus germplasm. Correlations between the variable pair reduce the evaluation effort and the resources to measure the genotype allocations in heterogeneous groups presenting high genetic variability. This makes it possible to select individuals for hybridization programs with F1 generation gains. Correlation and relative contribution networks, based on relationships graphical between fruit biometric characteristics, allow the variables selection with less effort and fewer measurements. O. distichus fruit biometric characters are efficient to quantify genetic divergence between genotypes.
Similar content being viewed by others
Introduction
The genetic diversity of Brazilian Amazonian plants is significant but remains poorly understood. The sites identification for conservation and germplasm collection in this region is a great challenge, especially for some native palm trees1.
Palm fruits and palm hearts native to the Amazon are consumed by local communities but are poorly known at the national and international levels2. Oenocarpus spp. plants have economic, ecological and dietary potential and are exploited through extraction by Amazonian communities1,3.
Six species of the genus Oenocarpus spp. are native to Brazil, but not endemic, including O. distichus Mart., O. bacaba Mart., O. minor Mart. and O. mapora H. Karten and O. pataua Mart.4. Oenocarpus distichus (also known as bacaba-de-leque) has high potential in the palm and processed pulp markets, including its use in the production of “bacaba wine”, a nutritious energy drink with commercial potential similar to that of Euterpe oleracea Mart.5.
Oenocarpus distichus phytochemicals have phenolic compounds correlated with antioxidant capacity2,6,7 and vitamin E, riboflavin, insoluble fiber, energy, and minerals, especially potassium and calcium8. This plant has oil with organoleptic properties similar to those of olive oil and raw material for the food industry with proteins of high biological value (40% more than soy)5 and adequate to manufacture ice cream and soap9.
The genetic diversity in O. distichus native populations, from the morphological characteristics of their fruits, needs to be better understood for germplasm inclusion. This is necessary for breeding programs to conserve native populations in extractive areas. Oenocarpus distichus genetic improvement and conservation programs depend on fruits biometric characterization and genetic parameter studies of genotypes showing promise for germplasm banks and conservation programs of this plant4,10.
Multivariate analyses allow selecting individuals for breeding programs and morphological or molecular characteristic identification to evaluate the divergence of native and exotic plants such as E. oleracea11, Theobroma grandiflorum Willd. Ex Spreng.12, Elaeis guineenses Jacq.13 and Phoenix dactylifera L.14 in forest fragments15. Grouping methods, based on prefixed similarity/dissimilarity measures, can identify divergent groups. Group determination, based on a standard, universally, accepted method does not exist, but individuals of the same group should be as homogeneous as possible and different from the others16.
Fruit characteristics are important for characterization, individual selection and genotypes choice during recombination cycles15,17. Genetic divergence, important for prospecting in plant breeding programs, allows us to know the available germplasm to evaluate similar or divergent groups to identify hybrid combinations with greater heterozygosity and heterotrophic effect18. Genetic divergence will allow us to know the available germplasm to predict combinations with higher heterozygosity so that there will be more possibilities of recovering fruitful Oenocarpus distichus genotypes in the segregant generations. Distribution among groups will allows the early determination of the best combinations and reduces the unneeded crosses number, prioritizing more contrasting groups for obtention of combination with higher heterosis. Biometric fruit variables differences such as morphological and production are regarded in the dissimilarity quantification. Those genetic variations among Oenocarpus distichus genotypes are essential for predicting heredity and level of heterosis, which are essential for improving Oenocarpus distichus production.
The objective of the present study was to quantify the genetic divergence between Oenocarpus distichus genotypes in a native population in the northeastern region of Pará, Amazon rainforest, Brazil, based on fruit biometric characteristics.
Results
All O. distichus fruit biometric characteristics (F-test < 0.01) show genetic variability (Table 1) with residual variation coefficients (CVe) of 6.67% for fruit length (FL) at 15.09% for pulp mass (PM), indicating high experimental precision. The genetic variation coefficient (CVg) values, mainly of the PM, seed mass (SM) and fruit mass (FM) characters, confirmed the genetic variation between the O. distichus genotypes. The relationship between CVg/CVe was greater than 1 for the characteristics fruit width (WF), FM, PM and SM. Matrix inheritance estimates for average ones (h2mp) ranged from 85.38 for pulp thickness (PT) to 96.81 for FM. The mean values of biometric characteristics of O. distichus fruits and Scott-Knott test at 5% probability are given in supplementary information.
The biometry correlation matrix of the O. distichus fruits indicate estimates for the last two eigenvalues near to zero, resulting in two linear relationships determining the harmful effects of multicollinearity (Table 2). The condition number for this matrix indicated NC > 1000, that is, severe colinearity. The eigenvectors, associated with the last two eigenvalues destacados, indicated that the FM and PM characteristics (highest absolute values highlighted in bold) are responsible for multicollinearity. The new NC matrix lower than 100, after the exclusion of the FM and PM variables, indicates weak collinearity (Table 3).
In addition to the CN, small single values were observed for FM (0.0503) and PM (0.0043) and high condition index FM (46.60) and PM (537.90), indicating serious problems caused by multicollinearity. VIF values can also be used to detect the existence of multicollinearity. The FM and PM variables presented VIFs of 12.53 and 101.37, respectively. The existence of at least one VIF with a value greater than 10 is sufficient for the regression coefficients associated with these values to be highly influenced by multicollinearity, indicating that these variables must be removed from the analyzes19.
The PM characteristic correlated with FL and PT and FM with AT and SM, all above 70% (Fig. 1B). The correlation pattern between the fruit biometric characteristics did not vary, even after excluding the FM and PM variables that caused multicollinearity (Fig. 1C,D). The characteristics PY and SM (Fig. 1C) and AT, SM, PT and WF (Fig. 1D) correlated, negatively, with a magnitude of 42% and above 70%, respectively.
Phenotypic correlation network of biometric characters fruit length (FL); fruit width (WF); fruit mass (FM); pulp mass (PM); seed mass (SM); pulp thickness (PT); almond thickness (AT) and pulp yield per fruit (PY) considering: (A,B) correlation network under the multicollinearity effect (NC > 1000) and (C,D) without multicollinearity effect (NC < 100), (A,C) correlation > 0.3 and (B,D) correlation > 0.7. Green lines mean positive correlation and negative redlines. The line thickness is proportional to the correlation intensity. Phenotypic correlation network generated with GENES SOFTWARE (Genes Software – extended and integrated with the R, Matlab and Selegen. http://arquivo.ufv.br/dbg/genes/genes_Br.htm).
Genetic distances between genotype pairs of O. distichus ranged from 0.07 to 48.10, with a mean of 11.44 (Table 4). Genotypes 3 and 15 formed the least divergent pair and 2 and 8 formed the most divergent one. The mean distances for 39% of genotype pairs were above the overall mean. The grouping of Tocher separated O. distichus genotypes into three genetically distinct groups (Table 5) with 80% of the genotypes in group I and a single palm in III (8), being the most divergent. The UPGMA method showed several groups, but a cut based20 reduced them to only three divergent genotype groups. The stratification in genetically different groups was equal to that of Tocher, with most of the genotypes in group I, and group III presenting only one genotypes at 8 (Fig. 2). The calculated cophenetic correlation coefficient (r = 0.96) in this study indicated that how similar the final hierarchical pattern and initial similarity (or distance) matrix are. Moreover, were obtained distortion and stress of 16 and 20%, respectively.
Dendrogram obtained by unweighted pair group method with arithmetic mean (UPGMA) based on Mahalanobis distance of fifteen Oenocarpus distichus genotypes with the cut-off point determined according to20 and cophenetic correlation coefficient (CCC). Dendrogram generated with GENES (GENES - a software package for analysis in experimental statistics and quantitative genetics. http://arquivo.ufv.br/dbg/genes/genes_Br.htm).
The first two canonical variables explained more than 80% of the total data variation (Table 6). The relative contribution of the fruit biometric variables to the genetic divergence was from 9.00 to 39.88% among the O. distichus palms and SM, AT and WF, with the greatest contribution to genetic divergence. The graphic dispersion in the two-dimensional space of 15 genotypes of O. distichus in relation to the first two canonical variables (Fig. 3), after exclusion of the least important variable, FL (CR = 9%), allowed the grouping of accessions similar to the optimization methods (Table 6) and UPGMA (Fig. 2).
Canonical variable 2D biplot related to six fruits traits of fifteen Oenocarpus distichus genotypes. Cophenetic correlation coefficient, distortion and stress of 96%, 16% and 20%, respectively. 2D biplot generated with GENES (GENES - a software package for analysis in experimental statistics and quantitative genetics. http://arquivo.ufv.br/dbg/genes/genes_Br.htm).
Discussion
The genetic variability between the O. distichus genotypes, for all fruit biometric characteristics, indicates that the germplasm of this plant can be collected in the region studied. Additionally, the selection, based on any of the eight features, provides genetic advantages to differentiate individuals within the same population, due to the cross between genetically different individuals causing greater heterotrophic effect21. The variability between the genotypes can be attributed to the data quantitative nature, polygenic, controlled by many genes13,22 and is important in ex situ genetic conservation, due to increasing the effective population size and reducing the mortality caused by inbreeding23. The efficiency of these biometric characteristics to distinguish genotypes from the family Arecaceae was confirmed with similar results for E. oleracea11,12,13,14,15,16,17,18,19,20,21. The CVg/CVe ratio (>1) and the high h²mp values, confirm the variability and the genetic control of fruit characteristics, allowing selecting and recombining the most promising O. distichus genotypes. CVg/CVe values, greater than 1, indicate gains in selection24 and that the environment has a low impact on the characteristics measured. These values should be considered in breeding and conservation programs for this palm10. Genetic variations in native populations are important to obtain parents to produce superior and divergent individuals for the interest characteristics in the F1 generation22,25,26. However, collection intensity may reduce fruit availability for frugivorous animals, compromising gene flow and genetic diversity in the forest fragment27. Therefore, conservation strategies such as not harvesting the fruits in years of low productivity and maintenance of at least 50% of them on the palms are important to maintain consumption by wildlife28.
The high multicollinearity values caused by FM and PM were expected, because they are characteristics collinear with PY and common in studies involving fruit characteristics, as reported for Theobroma grandiflorum Willd. Ex Spreng.12 and Plukenetia volubilis L.29. However, they should be excluded from the analyses, because they may lead to false biological interpretations and errors in the genotype selection process, due to the lack of precision in parameter estimation.
The correlations between the fruit biometric variables, visualized by graphic network analysis, show the potential of this technique to select variables that reduce the evaluation time and resources for measurement in O. distichus breeding programs. This confirms their wide use to relate characteristics of interest and to enable greater effectiveness in the divergent selection and superior O. distichus genotypes, as reported for Capsicum spp. Species30, Passiflora edulis Sims31 and Cocos nucifera L.22.
The large number of genotypes with genetic distances above the general average confirms genetic divergence and suggests the crosses’ ability to explore additive genetic values in O. distichus individuals and consequently their heterosis in subsequent cycles, as reported for P. volubilis29,32. Similar groupings with the Tocher and UPGMA methods, the consistent genetic divergences between genotypes 2, 5 and 8 and the low number of groups may indicate a common ancestry for this plant in the region12, similar to that reported for Bactris gasipaes Kunth.33, E. oleracea Mart.11 and T. grandiflorum Willd. Ex Spreng.12 with 3, 5 and 3 groups, respectively. Matrix separation into heterogeneous groups may indicate distinct gene pools for controlled hybridizations and reciprocal recurrent selection and, therefore, be interesting for genetic improvement18. Additionally, the grouping restricts the crosses number, reducing costs and increasing genetic gains in subsequent cycles, due to the reduced in breeding coefficient in the population by excluding similar individuals32.
The concentration of a large part of the total variance in the first two canonical variables (above 80%) shows that it is possible to study the genetic divergence between O. distichus genotypes by geometric distances in scatter plots34, as reported for genetic divergence in C. nucifera22. The projection of dissimilarity measures in 2D projection representation and use of dendrograms are acceptable when the correlation value between measures of original distances and a graphics is higher than 0.9 and values of distortion and stress lower than 20%35. In a graphic dispersion of canonical variables the grouping was compatible with the ones grouped by Tocher’s method. The bigger contribution of the SM, AT and WF characteristics to genetic divergence is important in selection programs, for being more responsive in the superior genotype identification32. The variable FL, even presenting lower relative contribution to genotype divergence, should not be removed from the analyses because it is easy to determine using non-destructive methods36. In addition, most of the genotypes that had lower FL showed higher yields of pulp (supplementary information). This result is in agreement with what is reported in the literature for E. oleracea, where higher fruit lengths result in lower pulp yields37.
Conclusions
Biometric fruit characteristics are efficient to quantify genetic divergence between Oenocarpus distichus genotypes. The contribution of the seed mass, almond thickness and fruit width, for genetic divergence, is higher and for this reason, these parameters should be prioritized in selection processes. Correlation networks facilitate the visualization of the correlations between the variables and the potential to choose those requiring less time for evaluation and fewer resources in the measurements in O. distichus breeding programs.
Grouping methods are effective in allocating O. distichus genotypes, in different groups, and should be used to maintain the genetic variability, conservation and selection of genotypes of this plant for breeding programs in native populations.
Material and Methods
Study area characterization
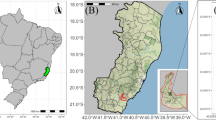
Fruits were collected from O. distichus native genotypes in an Amazonian forest fragment in the municipality of Oeiras do Pará, northeast of Pará, Brazil (02°00′11″S; 49°51′16″W) (Fig. 4). The climate is Ami type, according to Köppen’s classification, with 2.334 mm year−1 annual precipitation concentrated from December to March. The annual average temperature and relative air humidity is 83% and 29 °C, respectively38. The original forest types are dense low plateau forests and dense alluvial, with high populations of the genus Oenocarpus.
Geographic location of the municipality of Oeiras do Pará, Pará, Brazil. Map generated with ArcGis 10.6.1 (ESRI Development Team, 2018. ArcGis Geographic information system (GIS) for working with maps and geographic information. https://www.esri.com).
Plant material
Oenocarpus distichus genotypes, with mature fruits, were selected and those in good phytosanitary conditions were collected in April 2016, based on fruit productivity and with a minimum distance of 150 meters between genotypes. The fruits of each genotypes were identified, packed in plastic bags, placed in a Styrofoam box with ice and transported to the genetics and improvement laboratory of the Universidade Federal Rural da Amazônia, Campus Belém, Pará.
Biometric and statistical analysis
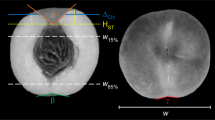
The statistical design was entirely random, with 15 genotypes and 14 fruits collected from each one. The length (FL), width (WF) (mm) and mass (FM) of fruits, pulp (PM) and seed (SM) (g) mass; and pulp (PT) and almond (AT) (mm) thickness were evaluated (Fig. 5). The pulp yield per fruit (PY) was obtained by the ratio between the FM and PM. The masses were determined on a digital scale (0.001 g) and the length and width using pachymeter (0.01 mm).
Genetic variability between genotypes was tested with analysis of variance (ANOVA using a Fisher’s test (F-test) at 1% probability. The genetic parameters heritability (h2mp), coefficient of genetic variation (CVg%) and variation coefficient (CVe%) were also determined. The means were grouped by Scott-Knott method39.
Possible linear relationships in the residual correlation matrices were obtained with multicollinearity test40. The multivariate analysis of O. distichus fruit data was performed using canonical variable techniques41. Dissimilarity was determined with the Mahalanobis generalized distance42 with grouping technique. The Tocher optimization method41 with the divergence measure mean per group lower than those between any other, and using the arithmetic mean method between unweighted pairs (UPGMA-Unweighted Pair Group Method with Arithmetic Average)43 based on Mojena20 delimited the groups. The relative contribution of the characteristics to genetic divergence was quantified by Mahalanobis generalized distances using the Singh method44.
Relationships between the fruit biometric evaluations were represented graphically with correlation networks. The phenotypic correlation were analyzed by weighted matrix with connections between the variables determined by the adjacent matrix A = h (R) as a function of: H(r ij) = ½ {sgn (│rij│−ρ) + 1}, in which ρ (hard limit) = 0, which allows viewing the connections. Network graphs were obtained with the variables with positive correlations connected by a green line and the negative ones by a red line. The line’s thickness represents the absolute correlation value with the thicker the line. The line’s thickness was controlled with a cut-off value of 0.3 and 0.7, meaning that only |rij| ≥ 0.3 and |rij| ≥ 0.7 have their lines highlighted, for easy viewing. The layout for the network was created with Fruchterman-Reingold algorithm45. The analyses were performed with Genes software46 and integration was realized with R47. The integration of the correlation network was evaluated with the “Qgraph” package48.
References
Moscoso, V., Albernaz, A. L. & Salomão, R. de. P. Niche modelling for twelve plant species (six timber species and six palm trees) in the Amazon region, using collection and field survey data. For. Ecol. and Manag. 310, 652–662, https://doi.org/10.1016/j.foreco.2013.08.064 (2013).
Sousa, S. H. B., Mattietto, R., de, A., Chisté, R. C. & Carvalho, A. V. Phenolic compounds are highly correlated to the antioxidant capacity of genotypes of Oenocarpus distichus Mart. fruits. Food Res. Inter. 108, 405–412, https://doi.org/10.1016/j.foodres.2018.03.056 (2018).
Carvalho, A. V., Silveira, T. F., Sousa, S. H. B., Moraes, M. R. & Godoy, H. T. Phenolic composition and antioxidant capacity of bacaba-de-leque (Oenocarpus distichus Mart.) genotypes. J. of F. Comp. and Anal. 54, 1–9, https://doi.org/10.1016/j.jfca.2016.09.013 (2016).
Moura, E. F., Oliveira, M., do, S. P., Silva, D. T. & Pontes, L. C. G. Genetic diversity and structure of Oenocarpus mapora germplasm conserved at eastern Amazon. Rev. Bras. de Frut. 37, 984–992, https://doi.org/10.1590/0100.2945-208/14 (2015).
Balick, M. J. Systematics and economic botany of the Oenocarpus-Jessenia (Palmae) complex. Adv. in Econ. Bot. 3, 1–140 Available from, http://www.nybgpress.org/Content/Site185/FilesSamples/183697AdvEcoBot_00000102745.pdf. (1986).
Canuto, G. A. B., Xavier, A. A. O., Neves, L. C. & Benassi, M. D. T. Caracterização físico-química de polpas de frutos da Amazônia e sua correlação com a atividade anti-radical livre. Rev. Bras. de Frut. 32, 1196–1205, https://doi.org/10.1590/S0100-29452010005000122 (2010).
Finco, F. D. B. A. et al. Antioxidant activity and characterization of phenolic compounds from bacaba (Oenocarpus bacaba Mart.) fruit by HPLC-DAD-MS. J. of Agri. and F. Chem. 60, 7665–7673, https://doi.org/10.1021/jf3007689 (2012).
Aguiar, J. P. L. & Souza, F. das C. do A. Bacaba-de-leque (Oenocarpus distichus): A new wet tropics nutritional source. Afri. J. of Agri. Res. 13, 803–805, https://doi.org/10.5897/AJAR2018.13027 (2018).
Clement, C. R., Lleras, E. & Van Leeuwen, J. O potencial das palmeiras tropicais no Brasil: acertos e fracassos das últimas décadas. Agrociencia 9, 67–71 Available from, http://fagro2.fagro.edu.uy/agrociencia/index.php/directorio/article/view/279/209 (2005).
Elameen, A. et al. Phenotypic diversity of plant morphological and root descriptor traits within a sweet potato, Ipomoea batatas (L.) Lam., germplasm collection from Tanzania. Gen. Res. and Crop Evol. 58, 397–407, https://doi.org/10.1007/s10722-010-9585-1 (2011).
Oliveira, M., do S. P., Ferreira, D. F., Santos, J. B. Divergência genética entre acessos de açaizeiro fundamentada em descritores morfoagronômicos. Pesq. Agro. Bras. 42, 501–506 Available from, http://www.scielo.br/pdf/%0D/pab/v42n4/07.pdf (2007).
Alves, R. M., Silva, C. R., de, S., Albuquerque, P. S. B. & Santos, V. S. Phenotypic and genotypic characterization and compatibility among genotypes to select elite clones of cupuassu. Acta Amazonica 47, 175–184, https://doi.org/10.1590/1809-4392201602104 (2017).
Arolu, I. W. et al. Genetic variability analysis and selection of pisifera palms for commercial production of high yielding and dwarf oil palm planting materials. Ind. Crops and Prod. 90, 135–141, https://doi.org/10.1016/j.indcrop.2016.06.006 (2016).
Ahmed, M. V. O. M. et al. Use of multivariate analysis to assess phenotypic diversity of date palm (Phoenix dactylifera L.) cultivars. Sci. Hortic. 127, 367–371, https://doi.org/10.1016/j.scienta.2010.11.011 (2011).
Moreira, A. F. P. et al. Genetic diversity, population structure and genetic parameters of fruit traits in Capsicum chinense. Sci. Hortic. 236, 1–9, https://doi.org/10.1016/j.scienta.2018.03.012 (2018).
Saha, A. & Das, S. Feature-weighted clustering with inner product induced norm based dissimilarity measures: an optimization perspective. Machine Learning 106, 951–992, https://doi.org/10.1007/s10994-016-5623-3 (2017).
Oliveira, J., da, S., Faleiro, F. G., Junqueira, N. T. V. & Viana, M. L. Genetic and morphoagronomic diversity of Passiflora spp. based on quantitative measurements of flowers and fruits. Rev. Bras. de Frut. 39, 1–7, https://doi.org/10.1590/0100-29452017003 (2017).
Domiciano, G. P., Alves, A. A., Laviola, B. G. & Conceição, L. D. H. C. S. Parâmetros genéticos e diversidade em progênies de macaúba com base em características morfológicas e fisiológicas. Ciência Rural 45, 1599–1605, https://doi.org/10.1590/0103-8478cr20140909 (2015).
Neter, J., Wasserman, W. & Kutner, M. H. Applied linear regression models. Richard D. Irwin Inc., Homewood. 547 p. (1983).
Mojena, R. Hierarchical grouping methods and stopping rules: An evaluation. The Comp. J. 20, 359–363, https://doi.org/10.1093/comjnl/20.4.359 (1977).
Sousa, A. M., Oliveira, M., do, S. P. & Farias Neto, J. T. Genetic divergence among white-type acai palm accessions based on morpho-agronomic characters. Pesq. Agro. Bras. 52, 751–760, https://doi.org/10.1590/s0100-204x2017000900007 (2017).
Sobral, K. M. B. et al. Genetic diversity assessment among tall coconut palm. Rev. Caatinga 31, 28–39, https://doi.org/10.1590/1983-21252018v31n104rc (2018).
Tambarussi, E. V., Boshier, D., Vencovsky, R., Freitas, M. L. M. & Sebbenn, A. M. Inbreeding depression from selfing and mating between relatives in the Neotropical tree Cariniana legalis Mart. Kuntze. Cons. Gen. 18, 225–234, https://doi.org/10.1007/s10592-016-0896-4 (2017).
Vencovsky, R. Herança quantitativa. In: Paterniani, E., Viegas, G. P. Melhoramento e produção do milho. 2. ed. Campinas: Fundação Cargill, p. 137–214 (1987).
Gomes Junior, R. A. et al. Evaluation of interspecific hybrids of palm oil reveals great genetic variability and potential selection gain. Ind. Crops and Prod. 52, 512–518, https://doi.org/10.1016/j.indcrop.2013.10.036 (2014).
Silva, A. V. C. D., Amorim, J. A. E., Vitória, M. F. D., Ledo, A. D. S. & Rabbani, A. R. C. Characterization of trees, fruits and genetic diversity in natural populations of mangaba. Ciência e Agrotec. 41, 255–262, https://doi.org/10.1590/1413-70542017413048416 (2017).
Novello, M. et al. Genetic conservation of a threatened Neotropical palm through community-management of fruits in agroforests and second-growth forests. For. Ecol. and Manag. 407, 200–209, https://doi.org/10.1016/j.foreco.2017.06.059 (2018).
Brancalion, P. H. S., Viani, R. A. G., Aronson, J., Rodrigues, R. R. & Nave, A. G. Improving planting stocks for the Brazilian Atlantic forest restoration through community based seed harvesting strategies. Rest. Ecol. 20, 704–711, https://doi.org/10.1111/j.1526-100X.2011.00839.x (2012).
Rodrigues, H. S. et al. Genetic diversity among accessions of sacha inchi (Plukenetiavolubilis) by phenotypic characteristics analysis. Acta Amazonica 48, 93–97, https://doi.org/10.1590/1809-4392201702591 (2018).
Silva, A. R. D., Rêgo, E. R. D., Pessoa, A. M. D. S. & Rêgo, M. M. D. Correlation network analysis between phenotypic and genotypic traits of chili pepper. Pesq. Agro. Bras. 51, 372–377, https://doi.org/10.1590/S0100-204X2016000400010 (2016).
Rosado, R. D. S. et al. Genetic divergence between passion fruit hybrids and reciprocals based on seedling emergence and vigor. J. of Seed Sci. 39, 417–425, https://doi.org/10.1590/2317-1545v39n4183293 (2017).
Silveira, G. F. et al. Genetic divergence among Passiflora cristalina vanderpl & zappi. genotypes based on flower and fruit characteristics. Rev. Bras. de Frut. 38, 1–8, https://doi.org/10.1590/0100-29452016491 (2016).
Martel, J. H. I., Ferraudo, A. S., Môro, J. R. & Perecin, D. Estatística multivariada na discriminação de raças amazônicas de pupunheiras (Bactris gasipaes Kunth) em Manaus (Brasil). Rev. Bras. de Frut. 25, 115–118, https://doi.org/10.1590/S0100-29452003000100033 (2003).
Cruz, C. D., Carneiro, P. C. de S. & Regazzi, A. J. Modelos Biométricos Aplicados ao Melhoramento Genético. 3. ed. Viçosa, M. G. Ediora UFV, 2: 668p. (2014).
Cruz, C. D, Ferreira, F. M. & Pessoni, L. A. Biometria aplicada ao estudo da diversidade genética. 1 ed. Visconde do Rio Branco, M. G. Editora Suprema Gráfica, 1: 620p. (2011).
Chagas, K., Alexandre, R. S., Schmildt, E. R., Bruckner, C. H. & Faleiro, G. F. Divergência genética em genótipos de maracujazeiro azedo, com base em características físicas e químicas dos frutos. Rev. Ciência Agro. 47, 524–531, https://doi.org/10.5935/1806-6690.20160063 (2016).
Farias Neto, J. T., Resende, M. D. V., Oliveira, M. & do, S. P. Seleção simultânea em progênies de açaizeiro irrigado para produção e peso de fruto. Rev. Bras. de Frut. 33, 532–539, https://doi.org/10.1590/s0100-29452011000200025 (2011).
Instituto Nacional de Meteorologia, INMET. Dados históricos. Available from, http://www.inmet.gov.br/portal [accessed 09 January 2018].
Scott, A. J. & Knott, M. A cluster analysis method for grouping means in the analysis of variance. Biometrics, 507–512, https://doi.org/10.2307/2529204 (1974).
Montgomery, D. C. & Peck, E. A. Introduction to linear regression analysis. New York: J. Wiley, 504p (1981).
Rao, C. R. Advanced statistical methods in biometric research. 2° Ed. 364–369 p.
Mahalanobis, P. C. (1936) Distância generalizada em estatística. Anais do Instituto Natural de Ciências 2, 49–55 (1952).
Sneath, P. H. & Sokal, R. R. Numerical taxonomy. The principles and practice of numerical classification, 573 p (1973).
Singh, D. The relative importance of characters affecting genetic divergence. Indian Journal of Genetics and Plant Breeding 41, 237–245 Available from, http://www.indianjournals.com/ijor.aspx?target=ijor:ijgpb&volume=41&issue=2&article=010 (1981).
Fruchterman, T. M. & Reingold, E. M. Graph drawing by force directed placement. Software: Practice and experience 21, 1129–1164, https://doi.org/10.1002/spe.4380211102 (1991).
Cruz, C. D. Genes: a software package for analysis in experimental statistics and quantitative genetics. Acta Sci. Agro. 35, 271–276, https://doi.org/10.4025/actasciagron.v35i3.21251 (2013).
Cruz, C. D. Genes Software-extended and integrated with the R, Matlab and Selegen. Acta Scientiarum. Agronomy 38, 547–552, https://doi.org/10.4025/actasciagron.v38i4.32629 (2016).
Epskamp, S., Cramer, A. O., Waldorp, L. J., Schmittmann, V. D. & Borsboom, D. qgraph: network visualizations of relationships in psychometric data. J. of Stat. Soft. 48, 1–18 Available from, http://www.webpages.uidaho.edu/stevel/504/Network%20Visualizations%20of%20Variables%20Relationships.pdf (2012).
Acknowledgements
The authors gratefully acknowledge Dr. Cosme Damiao Cruz, professor in the Department of General Biology (DBG) at the Federal University of Viçosa - Brazil, for his valuable assistance. Dr. Cruz provided excellent advice for statistical analyses and offered good suggestions about genetic diversity using GENES Software (https://www.facebook.com/GenesNews/). We also acknowledge Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) by granting scholarship to authors. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001.
Author information
Authors and Affiliations
Contributions
G.G.C.M. and M.T.A. de G. conceived and designed the elements of this study. R.S.A.S. and G.G.C.M. contributed to field samplings and analyses biometrics. R.D.S.R. and G.G.C.M. developed the statistical analyses and created all of the tables and figures presented in this manuscript. W.S.R. and A.C.P.N. contributed to reviewing and editing. The Manuscript was written mainly per G.G.C.M., T.G.V.M., R.D.S.R. and J.C.Z. although all authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mendes, G.G.C., de Gusmão, M.T.A., Martins, T.G.V. et al. Genetic divergence of native palms of Oenocarpus distichus considering biometric fruit variables. Sci Rep 9, 4943 (2019). https://doi.org/10.1038/s41598-019-41507-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41507-4
This article is cited by
-
Analysis of genetic divergence in Psidium cattleyanum Sabine accessions based on morphological fruit descriptors
Genetic Resources and Crop Evolution (2024)
-
Assessment of the genetic diversity of Eplingiella fruticosa (Salzm. ex Benth.) Harley & J.F.B.Pastore (Lamiaceae) based on morpho-agronomic markers and chemical composition of essential oils
Genetic Resources and Crop Evolution (2024)
-
Assessment of the phenotypic diversity in natural populations of Annona coriacea Mart.: implications for breeding
Genetic Resources and Crop Evolution (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.