Abstract
Although the prevalence of Intracytoplasmic sperm injection (ICSI) has increased year by year, there remains concern about the safety of these procedures because of reports of the increased risk for imprinting disorders. Previous research has demonstrated that gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. However, the epimutations in ICSI offspring after removing the effect of gonadotropin stimulation and the possibility that epimutations are reversible by developmental reprogramming has not been investigated. Our study is the first to investigate the effect of ICSI itself on methylation and exclude the effect of superovulation using the kidney tissues from the adult and old mice. We found reduced methylation and up-regulated expression of the imprinted genes, H19, Mest and Peg3, in adult ICSI mice, but the above alterations observed in adult mice were not detected in old ICSI mice. At the Snrpn DMR, methylation status was not altered in adult ICSI-derived mice, but hypermethylation and correlated down-regulated expression of Snrpn were observed in old mice. In conclusion, ICSI manipulation and early embryo culture resulted in alterations of methylation in differentially methylated region of H19, Mest, Peg3 and Snrpn, and the alterations were reprogrammed by developmental reprogramming.
Similar content being viewed by others
Introduction
Intracytoplasmic sperm injection (ICSI), one type of assisted reproductive technologies (ART), was firstly presented as a successful method over two decades and nowadays has become a widely-accepted medical treatment for male infertility. With the rising utilization of ICSI, several concerns of the progeny, therefore, have been raised. In theory, the ICSI technology is in the absence of natural selection of sperm; any structural damage to the oocyte produced by the injection or even the transfer of heterologous substances, such as culture solution, that would not been passed on to the progeny normally, might increase the potential for epigenetic errors, or epimutations. Meanwhile, gonadotropin stimulation, and in vitro culture of germ cell and resulting early embryo followed by transfer of embryo in the ICSI program, might also induce a higher risk of certain developmental abnormalities or disease phenotypes in offspring1.
Indeed, several studies indicated that children conceived by ICSI are at an increased risk for birth defects2,3,4,5 and imprinted diseases6,7,8,9. Hansen et al. compared 1138 infants born following the use of ART (301 infants with ICSI and 837 with in vitro fertilization) with 4000 naturally conceived infants, suggesting infants conceived with the use of ART had twice as high a risk of a major birth defect as naturally conceived infants10. Meanwhile, a nationwide German cohort study showed the relative risk of major malformation at birth was 1.44 in pregnancies established after ICSI, as compared with spontaneously conceived pregnancies11. Besides, systemic reviews suggested that children born following ICSI are at a higher risk of autism than the general population12. Nevertheless, the mechanisms behind the excess of malformation are still unclear. The parental genetic factors and the ICSI program itself are the possible explanations.
Germ cells and early embryos are unique, because they must undergo extensive epigenetic reprogramming to reestablish developmental potency during each generation13. Firstly, on every reproductive cycle, genomic imprints in the parental gametes are erased. Then, imprints in the immature germ cells of the developing embryo were reestablished according to their fate either as male or female gametes. Finally, imprints were maintained by both the preimplantation (when the rest of the genome is demethylated) and postimplantation development. Therefore, genomic imprints are dynamically changing during the developments of germ cell and embryo14. ICSI treatment, which is operated in the timing of in vitro culture of germ cell and fertilization and early embryo development, may interfere with the acquisition and maintaining of genomic imprints. The modification of imprinting center region (ICR) or differentially methylated region (DMR) during ICSI treatment were proofed to might contribute to birth defects or imprinted diseases15.
A possible excess of genitor-urinary defects, cardiovascular defects, gastrointestinal defects and nervous defects was shown in children conceived by ICSI16,17,18. Until now, the long-term effects of ICSI treatment have not been fully evaluated, especially in the adult and old ones, both because most individuals born through ICSI are still under 25-year-old and because research on potential effects in human or animal offspring is still ongoing. The more important thing is that, among the studies devoted to finding the mechanisms behind the association between diseases and ICSI treatment, most of them focus on the cardiovascular, gastrointestinal and nervous system defects by using placenta, liver, muscle and brain tissues1,19,20,21, few have focus on the urinary system by using kidney tissue.
Therefore, in our study, we examined kidney tissues, which were seldom been reported, from both the adult and the old mice. ICSI program includes four procedures, gonadotropin stimulation, manipulation and injection of occytes and culture followed by transfer of embryos. Then, which aspect or aspects directly contributes to the genesis of defects in the offspring? Previous observations suggested gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice1 and alteration of fatty acid metabolism20,22. To exclude the effect of gonadotropin stimulation, we analyzed ICSI-derived mice with mice derived by two-cell embryos transfer (TCET) instead of traditionally control mice which were derived by natural mating23. A key difference between ICSI and TCET mice is that manipulation and injection of oocytes and culture of early embryo during the ICSI program are removed in the TCET process and replaced by in vivo fertilization (Fig. 1). We focused on paternally imprinted gene H19, and maternally imprinted genes Snrpn, Mest, and Peg3, which were reported kidney disease related genes24,25,26,27,28, and measured the DNA methylation and expression of these genes at their DMRs. In this research, we tried to elucidate the underlying epigenetic mechanisms for these defects and provided evidences for the safety of ICSI.
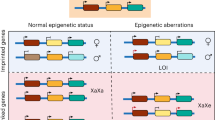
Schematic comparison of the TCET and ICSI procedures. (a) The TCET group is established by in vivo fertilization after gonadotropin stimulation. After 2-cell embryo is obtained from the oviduct, the embryo is subsequently transferred into a pseudo-pregnant mouse. (b) The ICSI mice also begin with the injection of gonadotropin and end with transferring 2-cell embryo into surrogate mice. A primary difference between the two procedures is the manipulation and injection of occytes and culture for ICSI, while only in vivo fertilization for TCET. (Drawn by authors Q. Z and X. Q).
Results
Alterations of methylation were detected in kidney tissues of adult ICSI mice
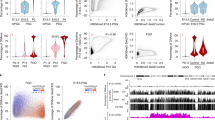
Kidney tissues from 16 ICSI-derived mice (8 adults and 8 olds) and 20 TCET-derived mice (10 adults and 10 olds) were used to analyze the DNA methylation and expression of the imprinted genes. DNA methylation level of imprinted gene H19 was first examined by Bisulfite sequencing PCR (BSP) in DMR island 3 containing 12 CpG sites. Compared with mice conceived by TCET, mice conceived through ICSI exhibited hypomethylation at all 12 CpG sites, ten of which (the 1st site and site from the 4th to the 12th) showed significantly hypomethylated level (p < 0.05) (Fig. 2d). Next, to verify the result of BSP, we used bisulfite pyrosequencing assay to examine 3 CpG sites of the region previously analyzed by BSP (Fig. 2a). Similar differential methylation was presented. Three CpG sites that showed relative hypomethylation in ICSI mice based on BSP also displayed reduced methylation when kidney tissues were examined by pyrosequencing (Fig. 3a). In addition, the 7th CpG site was significantly hypomethylated in ICSI mice (58.71% ± 1.49% in ICSI mice vs. 62.77% ± 1.56% in TCET mice, p < 0.01).
DNA methylation of the H19 and Snrpn DMRs in kidney tissues of adult and old ICSI-derived and TCET conceived mice. (a) Schematic representation of the differential methylated regions analyzed by bisulfite sequencing PCR (BSP) or pyrosequencing or both of them. The target sequences examined by pyrosequencing were in the boxes and CpG sites were underlined. (b) Methylation profiles by BSP (12 CpG sites) for the H19 DMR from ICSI-derived and TCET conceived mice. (c) Methylation profiles by BSP (16 CpG sites) for the Snrpn DMR from ICSI-derived and TCET conceived mice. (d) Statistical methylation analysis of H19 DMR and Snrpn DMR (Pearson chi-square test was used for the analysis: **p < 0.01; *p < 0.05).
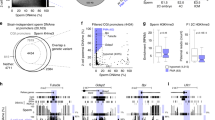
Pyrosequencing analyses of the methylation profiles from ICSI-derived, TCET conceived and natural mating mice of adult and old age. There are 3 CpG sites for the H19 DMR (a), 5 CpG sites for the Snrpn DMR (b), 4 CpG sites for the Mest DMR (c) and 5 CpG sites for the Peg3 DMR (d) (t-test was used for the analysis. ICSI versus TCET: **p < 0.01; *p < 0.05. TCET versus Nature: ##p < 0.01; #p < 0.05). The pyrogram of pyrosequencing of one sample in each gene and each group was shown in Supplementary Figure S1.
We used real-time RT PCR to quantify expression of H19 in kidney tissues from the same ICSI conceived and TCET-derived mice (Fig. 4a). Significantly up-regulated expressions were showed in ICSI mice, suggesting that the hypomethylation actually represented a valid change.
Maternally imprinted gene Snrpn in the same adult mice conceived either by ICSI or by TCET was also analyzed. We performed BSP to examine a position that contained 16 CpG sites in the Snrpn DMR. Over 80% (13 out of 16) CpG sites displayed reduced methylation in ICSI-derived mice when compared with TCET mice (Fig. 2d). The aberrant hypomethylation in ICSI mice also represented a valid alteration. There were two reasons: firstly, the mice derived by ICSI showed up-regulated expression compared with mice derived through TCET (Fig. 4a); secondly, the hypomethylation was also detected by bisulfite pyrosequencing (Fig. 3b).
DNA methylation at the second maternally imprinted gene Mest was also analyzed in the same ICSI mice and TCET mice. The DMR region containing 4 CpG sites was examined only by bisulfite pyrosequencing because of the consistent results of the previous genes obtained through BSP and pyrosequencing (Figs 2a,d and 3a and b). TCET mice exhibited differential methylation and the average methylated percentage was 60%. However, ICSI-derived mice displayed reduced methylation (on the 4th CpG sites: 60.57% ± 3.21% in ICIS mice vs. 67.00% ± 3.96% in TCET mice, p < 0.01) (Fig. 3c) and a correlated up-regulated expression of mRNA from the ICSI mice (Fig. 4a).
Finally, we analyzed the imprinted Peg3 gene, which is paternally expressed in kidney tissue. The region examined by pyrosequencing contained 5 CpG sites (Fig. 2a). TCET mice displayed differential methylation and the average methylation was 46%. While mice born through ICSI showed relative hypomethylation (on the 2nd CpG sites: 45.25% ± 2.49% in ICSI mice vs. 48.13% ± 2.80% in TCET mice, p < 0.05) (Fig. 3d) and also displayed a correlated up-regulated expression of mRNA compared with mice born through TCET (Fig. 4a).
Alterations of methylation might be reversible in kidney tissues of old ICSI mice
To determine whether methylation is dynamic during the development, we analyzed methylation and expression of the same imprinted genes in old mice born through ICSI and TCET. Surprisingly, we found hypermethylation at the maternally imprinted Snrpn DMR in the old ICSI-derived mice (p < 0.01) instead of hypomethylated methylation in the adult ICSI-derived mice when compared with the corresponding TCET mice (Fig. 2d). We also used bisulfite pyrosequencing assay to examine 5 CpG sites of the region previously analyzed by BSP. Similar differential methylation was presented. CpG sites that showed relative hypermethylation in ICSI old mice based on BSP also displayed a raised methylation when kidney tissues were examined by pyrosequencing (Fig. 3b). In addition, the 8th CpG site was significantly hypermethylated in ICSI mice (57.70%% ± 1.50% in ICSI mice vs. 54.10% ± 1.05% in TCET mice, p < 0.01).
The hypermethylation also represented a valid change, because a significantly down-regulated expression of Snrpn was shown in old ICSI mice (Fig. 4b). Nevertheless, methylation at the paternal imprinted H19 DMR, maternal imprinted Mest DMR and Peg3 DMR were examined, alterations of methylation that displayed in adult mice were not detected in old mice produced by ICSI compared with mice produced through TCET (Figs 2d and 3a,c,d). Similar expression of the above three imprinted genes H19, Mest, and Peg3, respectively, were also displayed in ICSI mice and TCET mice (Fig. 4b). Thus, it appears that DNA methylation can be reversible and always with correlated expression.
Hypermethylations were detected in kidney tissues of adult TCET mice and these alterations were reduced in kidney tissues of old TCET mice
De Waal E. et al. reported that gonadotropin stimulation contributed to an increased incidence of epimutations in ICSI-derived mice1. In our study, we evaluated the effects of gonadotropin stimulation and embryo transfer procedures by comparing the TCET mice and the natural mating (NM) mice. Hypermethylation was detected at the paternal imprinted H19 DMR, maternal imprinted Snrpn DMR, Mest DMR and Peg3 DMR (Fig. 3a–d). The 6th to the 8th CpG sites at H19 DMR were significantly hypermethylated in TCET mice compared with the NM control (the 6th CpG site: 53.56%% ± 1.51% in TCET mice vs. 50.25% ± 2.87% in NM mice, p < 0.05; the 7th CpG site: 62.78%% ± 1.56% in TCET mice vs. 59.13% ± 4.29% in NM mice, p < 0.05; the 8th CpG site: 54.78%% ± 4.84% in TCET mice vs. 48.75% ± 3.15% in NM mice, p < 0.01). Besides, raised methylations of the 2nd and the 5th CpG sites at Peg3 DMR were also shown significant (the 2th CpG site: 48.13%% ± 2.80% in TCET mice vs. 44.14% ± 1.46% in NM mice, p < 0.01; the 5th CpG site: 43.63%% ± 1.85% in TCET mice vs. 40.29% ± 1.80% in NM mice, p < 0.01).
Similarly, we analyzed methylation of the four imprinted genes in old mice born through TCET or conceived naturally. The significantly raised methylations that found in the adult TCET mice were not existed in the old TCET mice compared with the NM control (Fig. 3a–d). Therefore, alterations of DNA methylation produced by gonadotropin stimulation and embryo transfer procedures might be reduced during the development.
Discussion
Although the prevalence of ICSI has increased to about two-thirds or even higher of ART in some regions of the world29, there remains concern about the safety of this procedure because of reports of the increased risk for imprinting disorders. Of particular concern is the possibility that manipulations used in ICSI procedures might disrupt the normal acquisition and maintaining of genomic imprints during the development of germ cell and embryo. A systematic review of imprinting disorders in children conceived by ART demonstrated an increase in imprinted disorders, but evidence for an association between ART and methylation was still insufficient30. Studies in animals demonstrated that gonadotropin stimulation contributed to an increased incidence of epimutations in ICSI-derived mice1. However, few studies have reported the epimutations in ICSI offspring after removing the effect of gonadotropin stimulation. Besides, the possibility that epimutations are reversible by developmental reprogramming has not been investigated.
Our study is the first to investigate the effect of ICSI itself on methylation and exclude the effect of superovulation by using the kidney tissues from the adult and old mice. To exclude the effect of gonadotropin stimulation, we analyzed mice derived by TCET as control group (Fig. 1). Manipulation and injection of occytes and culture during the ICSI program were removed during the TCET process and replaced by in vivo fertilization. We found reduced methylation and up-regulated expression of imprinted genes in adult ICSI mice when compared with age-matched TCET mice, whereas in old ICSI-derived mice, hypermethylation at the Snrpn DMR and correlated down-regulated expression of Snrpn were observed when compared with the corresponding TCET mice. In addition, alterations of H19, Mest, and Peg3 methylation that observed in adult mice were not detected in old ICSI mice.
The effect of superovulation and embryo transfer procedures was also evaluated in kidney tissues. Hypermethyaltions were detected after the above procedures in the TCET offspring, which was consistent with the result of previous study1. And these alterations were reduced in the old mice, revealing that the hypermethylations might be corrected during the development. However, the TCET mice were surrogated by ICR mice instead of C57BL/6 J mice, so we could not eliminate the effect of maternal environment on the offspring.
Human studies reported higher methylation percentage of H19, Snrpn, Mest and Peg 3 in ART children than in naturally conceived children31–33. A meta-analysis of data from relevant reports revealed that the weighted mean difference (95 percentage confidence intervals) in percentage of methylation between IVF/ICSI versus spontaneously conceived were as follows: H19: −0.46(−1.41, 0.49), SNRPN: −0.55(−1.55, 0.46), MEST: −0.47(−2.07, 3.01) and PEG3: −0.24 (−1.72, 1.24), suggesting hyper-methylated DNA in ART IVF/ICSI children30. Our study excluded the effect of ovarian stimulation to the offspring, and founded hypo-methylated DNA in the ICSI adult when compared with TCET offspring, revealing the hypermethylation in ICSI children might be resulted from ovarian stimulation rather than ICSI manipulation itself.
Animal studies showed different effect of ART on different tissues, especially between the embryonic and extra-embryonic tissue. For instance, Fauque et al. showed that IVF and embryo culture resulted in hypomethylation of H19 DMR in placenta tissue, but not in fetal tissue34. Similarly, Mann et al. found loss of methylation at H19 DMR and Snrpn promoter in the placenta but not, or to a lesser extent, in fetus35. However, because of ethical question, human studies examined tissues easily obtained or relatively noninvasive. Tissues examined included peripheral blood, placenta, umbilical cord blood, buccal swabs and amnion/chorion. Thus, research about the effect of ART on different tissues was in desperate need.
Urogenital abnormality was one of the most common abnormalities of ART children, and H19, Mest, Peg3, and Snrpn were the related imprinting genes. In previous research, we had found that the methylation and expression of H19 in IVF offspring was changeable during the postnatal periods36. In this study, we found ICSI manipulation and early embryo culture really lead to methylation change of the above genes. More importantly, the methylation change was either reprogrammed or repaired during the development, and always with correlated mRNA expressions.
Methods
Animals
C57BL/6J female (6–8 weeks old) and male mice (8–10 weeks old) were used in this study to generate 16 mice conceived through the use of ICSI and 20 mice created through TCET. Kidney tissues were obtained from either adults (10 weeks old) or olds (1.5 years old), and then were used to analyze DNA methylation and expression of imprinted genes. For embryo transfer, both surrogated mothers (6–8 weeks old) and vasectomized males (8–10 weeks old) were ICR outbred mice.
Statements of approval and accordance
All methods were reviewed and approved by Zhejiang University Animal Care Committee, and all experiments were performed in accordance with the Institutional Guidelines for Animal Experiments (No. ZJU2009101007Y).
ICSI
ICSI was performed according to Wang and de Waal1,21. C57BL/6J female mice was subjected to hormone stimulation by consecutive injections of 7.5IU pregnant mareserum gonadotropin (PMSG, Organon) and 7.5 IU human CG (hCG, Organon) administered 46–48 hours apart. Approximately 13–15 hours after the injection of hCG, cumulus-oocyte complexes (COC) were collected from the oviducts. To remove cumulus cells, COC were treated with modified human tubal fluid medium (mHTF, Irvine). Cauda epididymal sperms of C57BL/6J male were suspended in warmed mHTF for 10 minutes, and a drop of sperm suspension was then mixed with polyvinylpyrrolidone (PVP, Irvine) in mHTF. The head of a single sperm was detached from the tail using a Piezo-driven pipette (PrimTech, Japan), and then injected into each oocyte by a micromanipulator (Narishige, Japan). Resulting zygotes were cultured for 24 hours to obtain two-cell stage embryos that were transferred into surrogate mice.
TCET
C57BL/6J female mice were caged with males of the same strain after stimulated by consecutive injections of PMSG and hCG as described in ICSI group. Female mice with a vaginal plug were separated from the males on day 2. Forty-four hours later, two-cell stage embryos were obtained from the oviducts and then were transferred into pseudo-pregnant surrogate mothers.
Embryo Transfer
All embryo transfers were performed with embryos at two-cell stage. Embryos derived from ICSI and TCET were transferred into the oviducts of ICR pseudo-pregnant mice (day 0.5) that had been mated with vasectomized ICR males the previous night. Newborn pups were obtained three weeks on average after embryo transfer. Sixteen mice conceived through the use of ICSI (8 adults and 8 olds) and 20 mice created through TCET (10 adults and 10 olds) were sacrificed to obtain kidney samples, which were then used to analyze the DNA methylation and expression of imprinted genes. Table 1 showed that body weights and weights of kidney tissues were similar between mice produced by TCET and mice conceived through ICSI (p > 0.05).
DNA Methylation Analysis
DNA was isolated from kidney tissue by using proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation (SangonBiotech) at all the time points.
Bisulfite sequencing PCR
Bisulfite sequencing PCR (BSP), which has served as the “gold standard” for measuring DNA methylation, was performed on genomic DNA (2 µg) by using the EpiTect Bisulfite kit (Qiagen) following the manufacturer’s instruction. Bisulfite-converted DNA (200 ng) was amplified by using PCR for H19 and Snrpn DMRs. MethPrimer software37 was used to identify CpG islands and to design primers. For the H19 DMR, the primer sequences were as follows: forward primer 5′-TTTTTGGGTAGTTTTTTTAGTTTTG-3′, reverse primer 5′-GTTTAATAAAGGGATTAGGTATTTGTGT-3′ (Wang et.al, 2012); and 12 CpG sites were analyzed (Fig. 2a). For the Snrpn DMR, the primer sequences were as follows: forward primer 5′-AATTTGTGTGATGTTTGTAATTATTTGG-3′, reverse primer 5′-TAAAATACACTTTCACTACTAAAATCC-3′; and 16 CpG sites were analyzed (Fig. 2a). The reaction conditions were as follows: predenaturation at 94 °C for 5 minutes, followed by 20 amplification cycles at 94 °C for 30 seconds, 60 °C to 50 °C (decreasing by 0.5 °C in each cycle) for 45 seconds, and 72 °C for 45 seconds, then a final extension and annealing step at 72 °C for 10 minutes21. Amplified products were gel purified by using NucleoSpin Extract II kit (Macherey-Nagel) according to the manufacture’s instruction. The purified products were ligated into the pMD 19-T Simple Vector (TaKaRa), and then transformed into Escherichia coli DH-5α competent cells and plated on LB plates with ampicillin (100 mg/mL) and incubated overnight at 37 °C. A single strain was inoculated into 2 mL LB liquid medium containing ampicillin (100 mg/mL) and grown overnight at 37 °C. Products were amplified using primer BcaBEST (TAKARA) which was specific for pMD 19-T Vector; and at least 10 clones from each sample for each imprinted genes with the expected band size were sequenced. The number of methylated CpGs (mCpGs) and unmethylated CpGs were recorded as previously described21. The percentage of overall mCpGs in each individual sites was calculated.
Bisulfite Pyrosequencing
To reduce the variability in the methylation of CpG sites by BSP that possibly due to the additional bacterial cloning step, we further assessed DNA methylation profiles of the H19 and Snrpn DMRs by using next-generation sequencing technology and performed custom pyrosequencing assays on 3 and 5 CpG sites, respectively, of the region previously analyzed by BSP (Fig. 2a). The results obtained through both methods were consistent (Results part). Because of the significant labor involved in BSP and the confirmed results of pyrosequencing, our study was limited to examining methylation of Mest and Peg3 DMRs only by pyrosequencing. The sequencing primer for bisulfate pyrosequencing was designed using Pyrosequencing Assay Design Software (Qiagen). Primer sequences are shown in Table 2. The conditions of amplification were the same as BSP. The biotinylated primer was used to facilitate extraction of single-stranded DNA templates by using streptavidin-coated Sepharose Beads (Amersham) according to manufacturer’s instruction. Biotinylated PCR product (10 µl) was used for pyrosequencing with the sequencing primer. For the H19 DMR, the sequencing primer was 5′-TTGTGTAGATTTGGTTATAG-3′ and 3CpG sites were analyzed. For the Snrpn DMR, the sequencing primer was 5′-TATGTGTAGTTATTGTTTGG-3′ and 5 CpG sites were analyzed. For the Mest DMR, the sequencing primer was 5′-GTAGTTTTTGGTATGTGGA-3′ and 4 CpG sites were analyzed. For the Peg3 DMR, the sequencing primer was 5′-ACCCAAAATAAACATCTCT-3′ and 5 CpG sites were analyzed (Fig. 2a). Bisulfite pyrosequencing was performed using the PyroMark Q24 ID System (Qiagen) and the PyroMark Gold Q24 reagents kit (Qiagen). The degree of methylation at each CpG site was quantified with Pyro Q-CpG software (Biotage) and determined by dividing the incorporation of cytosine by incorporation of cytosine plus thymine (%C = C/C + T) at each site. The percent methylation was calculated by averaging the degree of methylation at each of the CpG sites analyzed.
mRNA Expression of Imprinted Genes
RNA Extraction and Reverse Transcription (RT)
Total RNA was extracted from kidney tissues using a Trizol-based protocol. In brief, 10 mg kidney tissue was homogenized in 1 ml Trizol (Invitrogen). After adding chloroform, RNA was isolated from the aqueous phase according to the manufacturer’s instructions (Invitrogen). Isolated RNA (1 mg) was first treated with DNase (TaKaRa) in a 10-µl reaction with 5 × gDNA Eraser Buffer (2 µl) and gDNA Eraser (1 µl). The reaction was conducted at 42 °C for 2 minutes.
PrimeScript RT Reagent Kit (TaKaRa) was used for reverse transcription (RT) in a total volume of 20 µl with 4 µl 5 × PrimeScript Buffer, 1 µl PrimeScript RT Enzyme Mix I, 1 µl Oligo dT primer, and 1 µg of the above RNA sample. The mixture was incubated at 37 °C for 15 minutes, and the reaction was inactivated at 85 °C for 5 seconds.
Real-Time Quantitative PCR
PCR was performed by adding 1 µl cDNA (diluted five times before adding) to the PCR mix containing 5 µl 2 × SYBR Premix Ex Taq, 0.4 µl reverse primers and 0.2 µl ROX Reference Dye. It was carried out with one denature cycle at 95 °C for 10 seconds and 40 amplification cycles at 95 °C for 5 seconds and 60 °C for 30 seconds, followed by a dissociation curve which was constructed at 95 °C for 15 seconds, 60 °C for 15 seconds, and 75 °C for 15 seconds to detect the specificity of the PCR products, on the ABI 7900 real-time PCR system (Applied Biosystems) according to the manufacturer’s protocol. The mRNA levels of target genes were standardized with those of Gapdh reference gene and the average fold values were calculated. Primer sequences of PCR for these genes are shown in Table 3. Data (three biological repeats and three technical repeats at each time point) were analyzed by the comparative threshold cycle (CT) method and the standard formula38. Each reaction was performed in triplicate.
Statistical analysis
All computations were performed using SPSS 16.0. Comparisons between the two groups were made using unrelated t-tests and a Pearson chi-square test for BSP. The data are presented as the mean ± SD. In all cases, a values of p < 0.05 were considered statistically significant.
Data Availability
The datasets generated during or analyzed during the current study are available from the corresponding author on reasonable request.
References
de Waal, E. et al. Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Human Molecular Genetics. 21, 4460–4472 (2012).
Wen, S. W. et al. A comprehensive assessment of outcomes in pregnancies conceived by in vitro fertilization/intracytoplasmic sperm injection. Eur J Obstet Gynecol Reprod Biol. 150, 160–165 (2010).
Reefhuis, J. et al. Assisted reproductive technology and major structural birth defects in the United States dagger. Human Reproduction. 24, 360–366 (2009).
Farhi, A. et al. Congenital malformations in infants conceived following assisted reproductive technology in comparison with spontaneously conceived infants. J Matern FetalNeonatal Med. 26, 1171–1179 (2013).
Tararbit, K. et al. Risk of congenital heart defects associated with assisted reproductive technologies: a population-based evaluation. Eur Heart J. 32, 500–508 (2011).
Chang, A. S., Moley, K. H., Wangler, M., Feinberg, A. P. & DeBaun, M. R. Association between Beckwith-Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil. Steril. 83, 349–354 (2005).
Cox, G. F. et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am Soc Hum Genet. 71, 162–164 (2002).
Maher, E. R. et al. Beckwith-Wiedemannsyndrome and assisted reproduction technology (ART). J Med Genet. 40, 62–64 (2003).
Sutcliffe, A. G. et al. Assisted reproductive therapies and imprinting disorders—a preliminary British survey. Hum Reprod. 21, 1009–1011 (2006).
Hansen, M. et al. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. New England Journal of Medicine. 346, 725–730 (2002).
Katalinic, A. et al. Pregnancy course and outcome after intracytoplasmic sperm injection: a controlled, prospective cohort study. Fertility and Sterility. 81, 1604–1616 (2004).
Zhan, Q. T. et al. An overview of studies on psychological well-being in childrenborn following assisted reproductive technologies. J Zhejiang Univ-Sci B (Biomed & Biotechnol). 14, 947–960 (2013).
Morgan, H. D., Santos, F., Green, K., Dean, W. & Reik, W. Epigenetic reprogramming inmammals. Hum Mol Genet. 14, R47–R58 (2005).
Lucifero, D., Chaillet, J. R. & Trasler, J. M. Potential significance of genomic imprintingdefects for reproduction and assisted reproductive technology. Human Reproduction Update. 10, 3–18 (2004).
Rossignol, S. et al. The epigenetic imprinting defect of patients with Bechwith-Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet. 43, 902–907 (2006).
Zhu, J. L., Basso, O., Obel, C., Bille, C. & Olsen, J. Infertility, infertility treatment, and congenital malformations: Danish national birth cohort. BMJ. 333, 679 (2006).
Bonduelle, M. et al. A multi-centre cohort study of the physical health of 5-year-old children conceived after intracytoplasmic sperm injection, in vitro fertilization and natural conception. Hum Reprod. 20, 413–419 (2005).
Kurinczuk, J. & Bower, C. Birth defects in infants conceived by intracytoplasmic sperm injection: an alternative interpretation. Br Med J. 315, 1260–1265 (1997).
Sakian, S. et al. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta. 36, 1100–1105 (2015).
Wang, L. Y. et al. Superovulation Induced Changes of Lipid Metabolism in Ovaries and Embryos and Its Probable Mechanism. PLoSONE. 10, e0132638 (2015).
Wang, N. et al. Altered expressions and DNA methylation of imprinted genes in chromosome 7 in brain of mouse offspring conceived from in vitro maturation. Reproductive Toxicology. 34, 420–428 (2012).
Wang, L. Y. et al. Alteration of fatty acid metabolism in the liver, adipose tissue, and testis of male mice conceived through assisted reproductive technologies: fatty acid metabolism in ART mice. Lipids in Health and Disease. 12, 5 (2013).
de Waal, E. et al. Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. PNAS. 109, 4163–4168 (2012).
Riccio, A. et al. Inherited and Sporadic Epimutations at the IGF2-H19 locus in Beckwith-Wiedemann syndrome and Wilms’ tumor. Endocr Dev. 14, 1–9 (2009).
Dekel, B. et al. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 66, 6040–6049 (2006).
Kanwar, Y. S. et al. Imprinted mesodermal specific transcript (MEST) and H19 genes in renal development and diabetes. Kidney Int. 63, 1658–1670 (2003).
Schuster, A. E., Schneider, D. T., Fritsch, M. K., Grundy, P. & Perlman, E. J. Genetic and genetic expression analyses of clear cell sarcoma of the kidney. Lab Invest. 83, 1293–1299 (2003).
Frevel, M. A., Sowerby, S. J., Petersen, G. B. & Reeve, A. E. Methylation sequencing analysis refines the region of H19 epimutation in Wilms tumor. J Biol Chem. 274, 29331–29340 (1999).
Palermo, G. D., Neri, Q. V. & Rosenwaks, Z. To ICSI or not to ICSI. Semin Reprod Med. 33, 92–102 (2015).
Lazaraviciute, G. et al. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Human Reproduction Update. 21, 840–852 (2014).
Kanber, D., Buiting, K., Zeschnigk, M., Ludwig, M. & Horsthemke, B. Low frequency of imprinting defects in ICSI children born small for gestational age. Eur J Hum Genet. 17, 22–29 (2009).
Chan, W. E., Hatakeyama, C., Robinson, W. P. & Ma, S. DNA methylation at H19/IGF2 ICR1 in the placenta of pregnancies conceived by in vitro fertilization and intracytoplasmic sperm injection. Fertil Steril. 95, 2524–2526 (2011).
Rancourt, R. C., Harris, H. R. & Michels, K. B. Methylation levels at imprinting control regions are not altered with ovulation induction or in vitro fertilization in a birth cohort. Hum Reprod. 27, 2208–2216 (2012).
Fauque, P. et al. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Hum Mol Genet. 19, 1779–1790 (2010).
Mann, M. R. et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 131, 3727–3735 (2004).
Le, F. et al. In vitro fertilization alters growth and expression of Igf2/H19 and their epigenetic mechanisms in the liver and skeletal muscle of newborn and elder mice. Biology of Reproduction. 88, 570–578 (2013).
Li, L. C. & Dahiya, R. Meth Primer: designing primers for methylation PCRs. Bioinformatics. 18, 1427–1431 (2002).
Schefe, J. H., Lehmann, K. E., Buschmann, I. R., Unger, T. & Funke-Kaiser, H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. Journal of Molecular Medicine. 84, 901–910 (2006).
Acknowledgements
This work was supported by the National Basic Research Program of China (No. 2014CB943300; No. 2012CB944901), the National Natural Science Program of China (No. 81501321; No. 81200475), and the Zhejiang Provincial Natural Science Program of China (No. Y2100822; LY17H040006; LY14H040009; LQ14H040003; LZ13H040001).
Author information
Authors and Affiliations
Contributions
Q.Z., X.Q., and F.J. designed the experiments. N.W. and F.L. established the two animal models. Q.Z. and L.M. performed the bisulfite sequencing PCR and tissue analysis. Q.Z. performed the bisulfite pyrosequencing of the ICSI and TCET mice, and the work was assisted by X.Y., M.Y., H.L. and X.X. The bisulfite pyrosequencing of natural mating mice was finished by X.C. and X.Q. Data analysis and pictures used in the manuscript were finished by Q.Z. and X.Q. Manuscript was written by Q.Z. and modified by X.Q. and F.J.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhan, Q., Qi, X., Wang, N. et al. Altered methylations of H19, Snrpn, Mest and Peg3 are reversible by developmental reprogramming in kidney tissue of ICSI-derived mice. Sci Rep 7, 11936 (2017). https://doi.org/10.1038/s41598-017-11778-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-11778-w
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.