Abstract
Human pluripotent stem cells (hPSCs) hold a central role in studying human development, in disease modeling and in regenerative medicine. These cells not only acquire genetic modifications when kept in culture, but they may also harbor epigenetic aberrations, mainly involving parental imprinting and X-chromosome inactivation. Here we present a detailed bioinformatic protocol for detecting such aberrations using RNA sequencing data. We provide a pipeline designed to process and analyze RNA sequencing data for the identification of abnormal biallelic expression of imprinted genes, and thus detect loss of imprinting. Furthermore, we show how to differentiate among X-chromosome inactivation, full activation and aberrant erosion of X chromosome in female hPSCs. In addition to providing bioinformatic tools, we discuss the impact of such epigenetic variations in hPSCs on their utility for various purposes. This pipeline can be used by any user with basic understanding of the Linux command line. It is available on GitHub as a software container (https://github.com/Gal-Keshet/EpiTyping) and produces reliable results in 1–4 d.
Key points
-
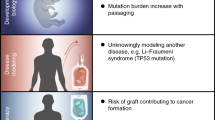
This protocol provides a step-by-step procedure to infer the presence of epigenetic aberrations in human pluripotent stem cells from RNA sequencing data, specifically focusing on the identification of imprinting and X-inactivation status.
-
The pipeline provides a simple and easy-to-use methodology for the reliable identification of gene- and locus-specific loss-of-imprinting events as well as the accurate discrimination among different X-inactivation statuses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The RNA-seq samples used as an example in this protocol, along with additional samples, can be retrieved from the SRA database. Their accession numbers are specified in Supplementary Table 2.
Code availability
All code used in this protocol is available at https://github.com/Gal-Keshet/EpiTyping and in the Supplementary Information.
References
De Los Angeles, A. et al. Hallmarks of pluripotency. Nature 525, 469–478 (2015).
Shahbazi, M. N., Siggia, E. D. & Zernicka-Goetz, M. Self-organization of stem cells into embryos: a window on early mammalian development. Science 364, 948–951 (2019).
Avior, Y., Sagi, I. & Benvenisty, N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 17, 170–182 (2016).
Trounson, A. & DeWitt, N. D. Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 17, 194–200 (2016).
Halliwell, J., Barbaric, I. & Andrews, P. W. Acquired genetic changes in human pluripotent stem cells: origins and consequences. Nat. Rev. Mol. Cell Biol. 21, 715–728 (2020).
Avior, Y., Lezmi, E., Eggan, K. & Benvenisty, N. Cancer-related mutations identified in primed human pluripotent stem cells. Cell Stem Cell 28, 10–11 (2021).
Lezmi, E. & Benvenisty, N. Identification of cancer-related mutations in human pluripotent stem cells using RNA-seq analysis. Nat. Protoc. 16, 4522–4537 (2021).
Ben-David, U., Mayshar, Y. & Benvenisty, N. Virtual karyotyping of pluripotent stem cells on the basis of their global gene expression profiles. Nat. Protoc. 8, 989–997 (2013).
Bar, S. & Benvenisty, N. Epigenetic aberrations in human pluripotent stem cells. EMBO J. 38, e101033 (2019).
Weinberger, L., Ayyash, M., Novershtern, N. & Hanna, J. H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17, 155–169 (2016).
Yilmaz, A. & Benvenisty, N. Defining human pluripotency. Cell Stem Cell 25, 9–22 (2019).
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21–32 (2001).
Tucci, V. et al. Genomic imprinting and physiological processes in mammals. Cell 176, 952–965 (2019).
Bar, S., Schachter, M., Eldar-Geva, T. & Benvenisty, N. Large-scale analysis of loss of imprinting in human pluripotent stem cells. Cell Rep. 19, 957–968 (2017).
Keshet, G. & Benvenisty, N. Large-scale analysis of imprinting in naive human pluripotent stem cells reveals recurrent aberrations and a potential link to FGF signaling. Stem Cell Rep. 16, 2520–2533 (2021).
Nora, E. P. & Heard, E. X chromosome inactivation: when dosage counts. Cell 139, 865–867 (2009).
Brown, C. J. et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542 (1992).
Heard, E. & Disteche, C. M. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 20, 1848–1867 (2006).
Shen, Y. et al. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc. Natl Acad. Sci. USA 105, 4709–4714 (2008).
Bruck, T. & Benvenisty, N. Meta-analysis of the heterogeneity of X chromosome inactivation in human pluripotent stem cells. Stem Cell Res. 6, 187–193 (2011).
Patel, S. et al. Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep. 18, 54–67 (2017).
Yokobayashi, S. et al. Inherent genomic properties underlie the epigenomic heterogeneity of human induced pluripotent stem cells. Cell Rep. 37, 109909 (2021).
Bar, S., Seaton, L. R., Weissbein, U., Eldar-Geva, T. & Benvenisty, N. Global characterization of X chromosome inactivation in human pluripotent stem cells. Cell Rep. 27, e3 (2019).
Werner, J. M., Ballouz, S., Hover, J. & Gillis, J. Variability of cross-tissue X-chromosome inactivation characterizes timing of human embryonic lineage specification events. Dev. Cell 57, 1995–2008.e5 (2022).
Theunissen, T. W. et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19, 502–515 (2016).
Sagi, I. & Benvenisty, N. Aspiring to naivety. Nature 540, 211–212 (2016).
Sarel-Gallily, R. & Benvenisty, N. Large-scale analysis of X inactivation variations between primed and naïve human embryonic stem cells. Cells 11, 1729 (2022).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Morison, I. M., Ramsay, J. P. & Spencer, H. G. A census of mammalian imprinting. Trends Genet. 21, 457–465 (2005).
Carrel, L. & Willard, H. F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005).
Tukiainen, T. et al. Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017).
Surani, M. A. H., Barton, S. C. & Norris, M. L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550 (1984).
McGrath, J. & Solter, D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 (1984).
Sagi, I. et al. Distinct imprinting signatures and biased differentiation of human androgenetic and parthenogenetic embryonic stem cells. Cell Stem Cell 25, 419–432.e9 (2019).
Cassidy, S. B., Schwartz, S., Miller, J. L. & Driscoll, D. J. Prader–Willi syndrome. Genet. Med. 14, 10–26 (2012).
Margolis, S. S., Sell, G. L., Zbinden, M. A. & Bird, L. M. Angelman syndrome. Neurotherapeutics 12, 641–650 (2015).
Weksberg, R., Shuman, C. & Beckwith, J. B. Beckwith–Wiedemann syndrome. Eur. J. Hum. Genet. 18, 8–14 (2010).
Ishida, M. New developments in Silver–Russell syndrome and implications for clinical practice. Epigenomics 8, 563–580 (2016).
Foong, Y. H., Thorvaldsen, J. L. & Bartolomei, M. S. Two sides of the Dlk1-Dio3 story in imprinting. Dev. Cell 56, 3035–3037 (2021).
Jinnah, H. A. Lesch–Nyhan disease: from mechanism to model and back again. Dis. Model. Mech. 2, 116–121 (2009).
Hoffman, E. P., Brown, R. H. & Kunkel, L. M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987).
Migeon, B. R. X-linked diseases: susceptible females. Genet. Med. 22, 1156–1174 (2020).
Mekhoubad, S. et al. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 10, 595–609 (2012).
Eisen, B. et al. Electrophysiological abnormalities in induced pluripotent stem cell‐derived cardiomyocytes generated from Duchenne muscular dystrophy patients. J. Cell. Mol. Med. 23, 2125 (2019).
Da Cruz, L. et al. Phase 1 clinical study of an embryonic stem cell-derived retinal pigment epithelium patch in age-related macular degeneration. Nat. Biotechnol. 36, 328–337 (2018).
Sonntag, K. C. et al. Pluripotent stem cell-based therapy for Parkinson’s disease: current status and future prospects. Prog. Neurobiol. 168, 1–20 (2018).
Chen, S., Du, K. & Zou, C. Current progress in stem cell therapy for type 1 diabetes mellitus. Stem Cell Res. Ther. 11, 275 (2020).
Lozano-Ureña, A. et al. Aberrations of genomic imprinting in glioblastoma formation. Front. Oncol. 11, 630482 (2021).
Fu, J. et al. DNA methylation of imprinted genes KCNQ1, KCNQ1OT1, and PHLDA2 in peripheral blood is associated with the risk of breast cancer. Cancers 14, 2652 (2022).
Zhou, J. et al. Epigenetic imprinting alterations as effective diagnostic biomarkers for early-stage lung cancer and small pulmonary nodules. Clin. Epigenetics 13, 220 (2021).
Lim, D. H. K. & Maher, E. R. Genomic imprinting syndromes and cancer. Adv. Genet. 70, 145–175 (2010).
Davies, H. D. et al. Myeloid leukemia in Prader–Willi syndrome. J. Pediatr. 142, 174–178 (2003).
Wang, D. et al. Abnormal X chromosome inactivation and tumor development. Cell. Mol. Life Sci. 77, 2949–2958 (2020).
Spatz, A., Borg, C. & Feunteun, J. X-chromosome genetics and human cancer. Nat. Rev. Cancer 4, 617–629 (2004).
Silva, S. S., Rowntree, R. K., Mekhoubad, S. & Lee, J. T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl Acad. Sci. USA 105, 4820–4825 (2008).
Bock, C. et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 28, 1106–1114 (2010).
Yong, W. S., Hsu, F. M. & Chen, P. Y. Profiling genome-wide DNA methylation. Epigenetics Chromatin 9, 26 (2016).
Irizarry, R. A. et al. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res. 18, 780–790 (2008).
Taiwo, O. et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc. 7, 617–636 (2012).
Brinkman, A. B. et al. Whole-genome DNA methylation profiling using MethylCap-seq. Methods 52, 232–236 (2010).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Li, Q., Hermanson, P. J. & Springer, N. M. Detection of DNA methylation by whole-genome bisulfite sequencing. Methods Mol. Biol. 1676, 185–196 (2018).
Kluin, R. J. C. et al. XenofilteR: computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinforma. 19, 366 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Brouard, J. S., Schenkel, F., Marete, A. & Bissonnette, N. The GATK joint genotyping workflow is appropriate for calling variants in RNA-seq experiments. J. Anim. Sci. Biotechnol. 10, 1–6 (2019).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Danecek, P. & McCarthy, S. A. BCFtools/csq: haplotype-aware variant consequences. Bioinformatics 33, 2037–2039 (2017).
Li, H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 27, 718 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Hu, Z. et al. Transient inhibition of mTOR in human pluripotent stem cells enables robust formation of mouse-human chimeric embryos. Sci. Adv. 6, eaaz0298 (2020).
Acknowledgements
We thank Assa Sherman for additional testing of the pipeline and providing constructive input, and all members of The Azrieli Center for Stem Cells and Genetic Research for critical reading of the manuscript. This work was partially supported by the Azrieli Foundation, the Rosetrees Trust, the US–Israel Binational Science Foundation (2021278), the Israel Science Foundation (2054/22), the ISF-Israel Precision Medicine Partnership (IPMP) Program (3605/21) and the HEAL project, funded by the European Union, EU Horizon (101056712). Views and opinions expressed are, however, those of the authors only and do not necessarily reflect those of the European Union or EU Horizon. Neither the European Union nor EU Horizon can be held responsible for them. N.B. is the Herbert Cohn Chair in Cancer Research.
Author information
Authors and Affiliations
Contributions
R.S.-G., G.K. and N.B. designed the analysis. R.S.-G. developed the X-inactivation analysis bioinformatic pipeline. G.K. developed the LOI analysis bioinformatic pipeline. R.S.-G. and G.K. wrote the manuscript and designed the automated code with input from S.K, G.H.-A. and N.B. N.B. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
N.B. is chief scientific officer of NewStem Ltd.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Bar, S. & Benvenisty, N. EMBO J. 38, e101033 (2019): https://doi.org/10.15252/embj.2018101033
Keshet, G. & Benvenisty, N. Stem Cell Rep. 16, 2520–2533 (2021): https://doi.org/10.1016/j.stemcr.2021.09.002
Sarel-Gallily, R. & Benvenisty, N. Cells 11, 1729 (2022): https://doi.org/10.3390/cells11111729
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Files 1–5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sarel-Gallily, R., Keshet, G., Kinreich, S. et al. EpiTyping: analysis of epigenetic aberrations in parental imprinting and X-chromosome inactivation using RNA-seq. Nat Protoc 18, 3881–3917 (2023). https://doi.org/10.1038/s41596-023-00898-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00898-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.