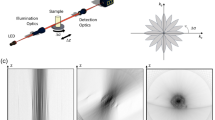
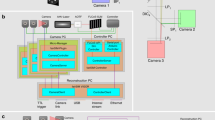
Abstract
With the development of a wide variety of animal models in recent years, there is a rapidly growing demand for long-term, high-speed intravital fluorescence imaging to observe intercellular and intracellular interactions in their native states. Scanning light-field microscopy (sLFM) with digital adaptive optics provides a compact computational solution by imaging the entire volume in a tomographic way with orders-of-magnitude improvement in spatiotemporal resolution and reduction in phototoxicity, as compared to traditional intravital microscopy. Here, we present a step-by-step protocol for both hardware and software implementation of multicolor sLFM as an add-on to a normal wide-field fluorescence microscope by using off-the-shelf lenses and devices at low cost. The procedure can be easily applied to other LFM variants, which can be advantageous in certain experimental contexts. Owing to the strong reliance of sLFM on algorithmic post-processing for high-quality data, the protocol describes various kinds of artefacts and corresponding parameters used for correcting and performance optimization. To increase the tolerance to system misalignment and differences in device fabrication, we describe a one-step calibration method for robust imaging performance up to the diffraction limit. An open-source graphical user interface is presented for hardware synchronization and real-time rendering of multiview images. The whole procedure including optical setup, software installation, system calibration and 3D reconstruction can be executed in 3–4 d with basic knowledge in optics and electronics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data supporting this study are available within the article, the Supplementary Information and the primary supporting study6. All data mentioned in the protocol are available in Supplementary Software.
Code availability
The Zemax source files and all code and software used in the protocol are available in Supplementary Software. We have also uploaded the code on GitHub (https://github.com/THU-IBCS/DAOSLIMIT_Protocol), which can be updated in the future with more functions.
References
Masedunskas, A., Porat-Shliom, N., Rechache, K., Aye, M.-P. & Weigert, R. Intravital microscopy reveals differences in the kinetics of endocytic pathways between cell cultures and live animals. Cells 1, 1121–1132 (2012).
Pittet, M. J., Garris, C. S., Arlauckas, S. P. & Weissleder, R. Recording the wild lives of immune cells. Sci. Immunol. 3, eaaq0491 (2018).
Winter, P. W. & Shroff, H. Faster fluorescence microscopy: advances in high speed biological imaging. Curr. Opin. Chem. Biol. 20, 46–53 (2014).
Liu, T. L. et al. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science 360, eaaq1392 (2018).
Icha, J., Weber, M., Waters, J. C. & Norden, C. Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays 39, 1700003 (2017).
Wu, J. et al. Iterative tomography with digital adaptive optics permits hour-long intravital observation of 3D subcellular dynamics at millisecond scale. Cell 184, 3318–3332.e17 (2021).
Levoy, M., Ng, R., Adams, A., Footer, M. & Horowitz, M. Light field microscopy. ACM Trans. Graph. 25, 924–934 (2006).
Prevedel, R. et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat. Methods 11, 727–730 (2014).
Cong, L. et al. Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio). Elife 6, e28158 (2017).
Nöbauer, T. et al. Video rate volumetric Ca2+ imaging across cortex using seeded iterative demixing (SID) microscopy. Nat. Methods 14, 811–818 (2017).
Pégard, N. C. et al. Compressive light-field microscopy for 3D neural activity recording. Optica 3, 517–524 (2016).
Yoon, Y.-G. et al. Sparse decomposition light-field microscopy for high speed imaging of neuronal activity. Optica 7, 1457–1468 (2020).
Broxton, M. et al. Wave optics theory and 3-D deconvolution for the light field microscope. Opt. Express 21, 25418–25439 (2013).
Taylor, M. A., Nöbauer, T., Pernia-Andrade, A., Schlumm, F. & Vaziri, A. Brain-wide 3D light-field imaging of neuronal activity with speckle-enhanced resolution. Optica 5, 345–353 (2018).
Hua, X., Liu, W. & Jia, S. High-resolution Fourier light-field microscopy for volumetric multi-color live-cell imaging. Optica 8, 614–620 (2021).
Wagner, N. et al. Instantaneous isotropic volumetric imaging of fast biological processes. Nat. Methods 16, 497–500 (2019).
Truong, T. V. et al. High-contrast, synchronous volumetric imaging with selective volume illumination microscopy. Commun. Biol. 3, 1–8 (2020).
Zhang, Z. et al. Imaging volumetric dynamics at high speed in mouse and zebrafish brain with confocal light field microscopy. Nat. Biotechnol. 39, 74–83 (2021).
Fu, Z. et al. Light field microscopy based on structured light illumination. Opt. Lett. 46, 3424–3427 (2021).
Zhang, Y. et al. Computational optical sectioning with an incoherent multiscale scattering model for light-field microscopy. Nat. Commun. 12, 6391 (2021).
Lin, X., Wu, J. & Dai, Q. Camera array based light field microscopy. Biomed. Opt. Express 6, 3179–3189 (2015).
Guo, C., Liu, W., Hua, X., Li, H. & Jia, S. Fourier light-field microscopy. Opt. Express 27, 25573–25594 (2019).
Lu, Z. et al. Phase-space deconvolution for light field microscopy. Opt. Express 27, 18131–18145 (2019).
Zhang, Y. et al. DiLFM: an artifact-suppressed and noise-robust light-field microscopy through dictionary learning. Light Sci. Appl. 10, 152 (2021).
Wang, Z. et al. Real-time volumetric reconstruction of biological dynamics with light-field microscopy and deep learning. Nat. Methods 18, 551–556 (2021).
Wagner, N. et al. Deep learning-enhanced light-field imaging with continuous validation. Nat. Methods 18, 557–563 (2021).
Alonso, M. A. Wigner functions in optics: describing beams as ray bundles and pulses as particle ensembles. Adv. Opt. Photonics 3, 272–365 (2011).
Zhu, S., Lai, A., Eaton, K., Jin, P. & Gao, L. On the fundamental comparison between unfocused and focused light field cameras. Appl. Opt. 57, A1–A11 (2018).
Abrahamsson, S. et al. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods 10, 60–63 (2013).
Nakano, A. Spinning-disk confocal microscopy—a cutting-edge tool for imaging of membrane traffic. Cell Struct. Funct. 27, 349–355 (2002).
Zipfel, W. R., Williams, R. M. & Webb, W. W. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 21, 1369–1377 (2003).
Bouchard, M. B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat. Photonics 9, 113–119 (2015).
Voleti, V. et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods 16, 1054–1062 (2019).
Fahrbach, F. O., Simon, P. & Rohrbach, A. Microscopy with self-reconstructing beams. Nat. Photonics 4, 780–785 (2010).
Ji, N. Adaptive optical fluorescence microscopy. Nat. Methods 14, 374–380 (2017).
Park, J. H., Kong, L., Zhou, Y. & Cui, M. Large-field-of-view imaging by multi-pupil adaptive optics. Nat. Methods 14, 581–583 (2017).
Lin, Q. et al. Cerebellar neurodynamics predict decision timing and outcome on the single-trial level. Cell 180, 536–551 (2020).
Quicke, P. et al. Subcellular resolution three-dimensional light-field imaging with genetically encoded voltage indicators. Neurophotonics 7, 035006 (2020).
Schoonover, C. E., Ohashi, S. N., Axel, R. & Fink, A. J. P. Representational drift in primary olfactory cortex. Nature 594, 541–546 (2021).
Plaçais, P. Y. et al. Upregulated energy metabolism in the Drosophila mushroom body is the trigger for long-term memory. Nat. Commun. 8, 1–14 (2017).
Jiang, D. et al. Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat. Cell Biol. 21, 966–977 (2019).
Ma, L. et al. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 25, 24–38 (2015).
Zheng, G., Horstmeyer, R. & Yang, C. Wide-field, high-resolution Fourier ptychographic microscopy. Nat. Photonics 7, 739–745 (2013).
Humphry, M. J., Kraus, B., Hurst, A. C., Maiden, A. M. & Rodenburg, J. M. Ptychographic electron microscopy using high-angle dark-field scattering for sub-nanometre resolution imaging. Nat. Commun. 3, 1–7 (2012).
Milkie, D. E., Betzig, E. & Ji, N. Pupil-segmentation-based adaptive optical microscopy with full-pupil illumination. Opt. Lett. 36, 4206–4208 (2011).
Boyd, S., Parikh, N., Chu, E., Peleato, B. & Eckstein, J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Found. Trends Mach. Learn. 3, 1–122 (2010).
Dabov, K., Foi, A., Katkovnik, V. & Egiazarian, K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans. Image Process. 16, 2080–2095 (2007).
Zhong, Q. et al. High-definition imaging using line-illumination modulation microscopy. Nat. Methods 18, 309–315 (2021).
Li, X. et al. Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising. Nat. Methods 18, 1395–1400 (2021).
Zhu, S., Tian, R., Antaris, A. L., Chen, X. & Dai, H. Near-infrared-II molecular dyes for cancer imaging and surgery. Adv. Mater. 31, e1900321 (2019).
Qian, Y. et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat. Methods 16, 171–174 (2019).
Nieuwenhuizen, R. P. J. et al. Measuring image resolution in optical nanoscopy. Nat. Methods 10, 557–562 (2013).
Acknowledgements
We thank D. Jiang and L. Yu for their assistance in sample preparation. We thank T. Zhu and T. Yan for insightful discussions. This work was supported by the National Natural Science Foundation of China (62088102 and 62071272) and the National Key Research and Development Program of China (2020AA0105500 and 2020AAA0130000). We further acknowledge support from the Beijing Laboratory of Brain and Cognitive Intelligence, the Beijing Municipal Education Commission and the Beijing Key Laboratory of Multi-dimension & Multi-scale Computational Photography (MMCP).
Author information
Authors and Affiliations
Contributions
Q.D., J.W. and Z.L. conceived the project. Z.L. and J.W. designed the whole pipeline. Z.L. and Y.C. conducted numerical simulations and biological experiments. Z.L. and Y.N. worked on data processing. J.W., Z.L. and Y.N. designed the Zemax and CAD prescriptions. Z.L. and Y.Y. prepared the acquisition and other pre-processing code for the protocol. Z.L, Y.C., J.W. and Q.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Q.D. and J.W. are co-founders and equity holders of Zhejiang Hehu Technology LLC, where the DAOSLIMIT technology is commercialized. Q.D., J.W. and Z.L. have patents related to the DAOSLIMIT technology (US Patent, no. 11,131,841).
Peer review
Peer review information
Nature Protocols thanks Robert Prevedel and Yicong Wu for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Wu, J. et al. Cell 184, 3318–3332.e17 (2021): https://doi.org/10.1016/j.cell.2021.04.029
Zhang, Y. et al. Nat. Commun. 12, 6391 (2021): https://doi.org/10.1038/s41467-021-26730-w
Zhang, Y. et al. Light Sci. Appl. 10, 152 (2021): https://doi.org/10.1038/s41377-021-00587-6
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, Supplementary Methods and Supplementary Manuals 1 and 2.
Supplementary Software
The DAOSLIMIT software package contains the GUIs, related codes, reconstruction codes, Zemax files and example raw data.
Rights and permissions
About this article
Cite this article
Lu, Z., Cai, Y., Nie, Y. et al. A practical guide to scanning light-field microscopy with digital adaptive optics. Nat Protoc 17, 1953–1979 (2022). https://doi.org/10.1038/s41596-022-00703-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00703-9
This article is cited by
-
Long-term intravital subcellular imaging with confocal scanning light-field microscopy
Nature Biotechnology (2024)
-
Virtual-scanning light-field microscopy for robust snapshot high-resolution volumetric imaging
Nature Methods (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.