Abstract
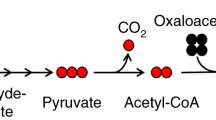
Targeted tandem mass spectrometry (LC–MS/MS) has been extremely useful for profiling small molecules extracted from biological sources, such as cells, bodily fluids and tissues. Here, we present a protocol for analysing incorporation of the non-radioactive stable isotopes carbon-13 (13C) and nitrogen-15 (15N) into polar metabolites in central carbon metabolism and related pathways. Our platform utilizes selected reaction monitoring (SRM) with polarity switching and amide hydrophilic interaction liquid chromatography (HILIC) to capture transitions for carbon and nitrogen incorporation into selected metabolites using a hybrid triple quadrupole (QQQ) mass spectrometer. This protocol represents an extension of a previously published protocol for targeted metabolomics of unlabeled species and has been used extensively in tracing the metabolism of nutrients such as 13C-labeled glucose, 13C-glutamine and 15N-glutamine in a variety of biological settings (e.g., cell culture experiments and in vivo mouse labelling via i.p. injection). SRM signals are integrated to produce an array of peak areas for each labelling form that serve as the output for further analysis. The processed data are then used to obtain the degree and distribution of labelling of the targeted molecules (termed fluxomics). Each method can be customized on the basis of known unlabeled Q1/Q3 SRM transitions and adjusted to account for the corresponding 13C or 15N incorporation. The entire procedure takes ~6–7 h for a single sample from experimental labelling and metabolite extraction to peak integration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Jorda, J. et al. Quantitative metabolomics and instationary 13C-metabolic flux analysis reveals impact of recombinant protein production on trehalose and energy metabolism in Pichia pastoris. Metabolites 4, 281–299 (2014).
Metallo, C. M., Walther, J. L. & Stephanopoulos, G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J. Biotechnol. 144, 167–174 (2009).
Nissim, I. et al. Effects of a glucokinase activator on hepatic intermediary metabolism: study with 13C-isotopomer-based metabolomics. Biochem. J. 444, 537–551 (2012).
Zamboni, N. & Sauer, U. Novel biological insights through metabolomics and 13C-flux analysis. Curr. Opin. Microbiol. 12, 553–558 (2009).
Wills, J., Edwards-Hicks, J. & Finch, A. J. AssayR: a simple mass spectrometry software tool for targeted metabolic and stable isotope tracer analyses. Anal. Chem. 89, 9616–9619 (2017).
Zamboni, N., Saghatelian, A. & Patti, G. J. Defining the metabolome: size, flux, and regulation. Mol. Cell 58, 699–706 (2015).
Zamboni, N., Fendt, S.-M., Rühl, M. & Sauer, U. 13C-based metabolic flux analysis. Nat. Protoc. 4, 878–892 (2009).
Nanchen, A., Fuhrer, T. & Sauer, U. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol. 358, 177–197 (2007).
Crown, S. B., Long, C. P. & Antoniewicz, M. R. Optimal tracers for parallel labeling experiments and (13)C metabolic flux analysis: a new precision and synergy scoring system. Metab. Eng. 38, 10–18 (2016).
Wiechert, W. 13C metabolic flux analysis. Metab. Eng. 3, 195–206 (2001).
Wiechert, W., Mollney, M., Petersen, S. & de Graaf, A. A. A universal framework for 13C metabolic flux analysis. Metab. Eng. 3, 265–283 (2001).
Weitzel, M. et al. 13CFLUX2--high-performance software suite for (13)C-metabolic flux analysis. Bioinformatics 29, 143–145 (2013).
Huang, X. et al. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal. Chem. 86, 1632–1639 (2014).
Noh, K., Droste, P. & Wiechert, W. Visual workflows for 13C-metabolic flux analysis. Bioinformatics 31, 346–354 (2015).
Yuan, M., Breitkopf, S. B., Yang, X. & Asara, J. M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7, 872–881 (2012).
Ying, H. et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012).
Ben-Sahra, I., Howell, J. J., Asara, J. M. & Manning, B. D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328 (2013).
Ben-Sahra, I., Hoxhaj, G., Ricoult, S. J. H., Asara, J. M. & Manning, B. D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016).
Shyh-Chang, N. et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 (2013).
Son, J. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013).
Nicolay, B. N. et al. Loss of RBF1 changes glutamine catabolism. Genes Dev. 27, 182–196 (2013).
Cox, A. G. et al. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat. Cell Biol. 18, 886–896 (2016).
Locasale, J. W. et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43, 869–874 (2011).
Hu, H. et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164, 433–446 (2016).
Lien, E. C. et al. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat. Cell Biol. 18, 572–578 (2016).
Sousa, C. M. et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016).
Viale, A. et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014).
Breitkopf, S. B., Taveira, M. O., Yuan, M., Wulf, G. M. & Asara, J. M. Serial-omics of P53–/–, Brca1–/– mouse breast tumor and normal mammary gland. Sci. Rep. 7, 14503 (2017).
Breitkopf, S. B. et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 13, 30 (2017).
Breitkopf, S. B., Yuan, M., Helenius, K. P., Lyssiotis, C. A. & Asara, J. M. Triomics analysis of imatinib-treated myeloma cells connects kinase inhibition to RNA processing and decreased lipid biosynthesis. Anal. Chem. 87, 10995–11006 (2015).
Hensley, C. T. et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016).
Davidson, S. M. et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016).
Lane, A.N., Yan, J. & Fan, T.W. 13C Tracer studies of metabolism in mouse tumor xenografts. Bio Protoc. 5, e1650 (2015).
Mashimo, T. et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159, 1603–1614 (2014).
Glick, G. D. et al. Anaplerotic metabolism of alloreactive T cells provides a metabolic approach to treat graft-versus-host disease. J. Pharmacol. Exp. Ther. 351, 298–307 (2014).
Juvekar, A. et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc. Natl. Acad. Sci. USA 113, E4338–E4347 (2016).
Huang, H., Yuan, M., Wulf, G. M. & Asara, J. M. in Proceedings from the ASMS Conference on Mass Spectrometry and Allied Topics (San Diego, CA, 2018).
Wang, J. B. et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18, 207–219 (2010).
Acknowledgements
This research was supported by grants from the National Institutes of Health (5P01CA120964 to J.M.A. and B.D.M.; 5R35CA197459 to B.D.M. and J.M.A.; and 5P30CA006516 to J.M.A.) and the BIDMC Research Capital Fund for funding the mass spectrometry instrumentation. C.A.L. was supported by a Pancreatic Cancer Action Network/AACR Pathway to Leadership award (13-70-25-LYSS), a Dale F. Frey Award for Breakthrough Scientists from the Damon Runyon Cancer Research Foundation (DFS-09-14), a Junior Scholar Award from The V Foundation for Cancer Research (V2016-009), a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research (SKF-16-005), and a 2017 AACR NextGen Grant for Transformative Cancer Research (17-20-01-LYSS). D.M.K was supported by a University of Michigan Program in Chemical Biology Graduate Assistance in Areas of National Need (GAANN) award. We thank L.C. Cantley (Weill-Cornell MC) and G.M. Wulf (BIDMC/HMS) for helpful discussions and the donation of K14 cells.
Author information
Authors and Affiliations
Contributions
J.M.A. developed the platform, incorporated the methods, analysed data and wrote the protocol. C.A.L. and D.M.K. helped to develop the SRM transitions for labeled metabolites, generated samples for testing the protocol, analysed data and edited the protocol. B.D.M. and I.B.S. helped to generate the SRM transitions for labeled metabolites, generated samples for testing the protocol and edited the protocol. M.Y. helped to incorporate methods, maintained the platform and helped with sample generation and data analysis. H.H. helped to develop the SRM transitions for labeled metabolites, generated samples for testing the protocol and helped with data analysis. S.B.B. incorporated methods and generated labeled samples for testing the protocol.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Protocol to which this paper is an extension
Yuan, M., Breitkopf, S. B., Yang, X. & Asara, J. M. Nat. Protoc. 7, 872–881 (2012) https://www.nature.com/articles/nprot.2012.024
Key references using this protocol
Son, J. et al. Nature 496, 101–105 (2013): http://www.nature.com/articles/nature12040
Ben-Sahra, I., Howell, J. J., Asara, J. M. & Manning, B. D. Science 339, 1323–1328 (2013): http://science.sciencemag.org/content/339/6125/1323
Ying, H. et al. Cell 149, 656–670 (2012): https://www.cell.com/cell/abstract/S0092-8674(12)00352-2
Ben-Sahra, I., Hoxhaj, G., Ricoult, S. J. H., Asara, J. M. & Manning, B. D.: Science 351, 728–733 (2016) http://science.sciencemag.org/content/351/6274/728
This protocol is an extension to: Yuan, M. et al., Nat. Protoc. 7, 872–881 (2012), doi:10.1038/nprot.2012.024; published online 12 April 2012.
Supplementary information
Supplementary Dataset 1
Supplementary Dataset 1
Supplementary Dataset 2
Supplementary Dataset 2
Supplementary Dataset 3
Supplementary Dataset 3
Supplementary Dataset 4
Supplementary Dataset 4
Supplementary Dataset 5
Supplementary Dataset 5
Rights and permissions
About this article
Cite this article
Yuan, M., Kremer, D.M., Huang, H. et al. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC–MS/MS. Nat Protoc 14, 313–330 (2019). https://doi.org/10.1038/s41596-018-0102-x
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0102-x
This article is cited by
-
Lactate-dependent transcriptional regulation controls mammalian eye morphogenesis
Nature Communications (2023)
-
Metabolic pathway analysis using stable isotopes in patients with cancer
Nature Reviews Cancer (2023)
-
Methionine restriction constrains lipoylation and activates mitochondria for nitrogenic synthesis of amino acids
Nature Communications (2023)
-
DDX39B facilitates the malignant progression of hepatocellular carcinoma via activation of SREBP1-mediated de novo lipid synthesis
Cellular Oncology (2023)
-
Uridine-derived ribose fuels glucose-restricted pancreatic cancer
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.