Abstract
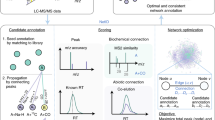
Identification of previously unreported metabolites (so-called ‘unknowns’) in untargeted metabolomic data has become an increasingly active area of research. Considerably less attention, however, has been dedicated to identifying unknown metabolic pathways. Yet, for each unknown metabolite structure, there is potentially a yet-to-be-discovered chemical transformation. Elucidating these biochemical connections is essential to advancing our knowledge of cellular metabolism and can be achieved by tracking an isotopically labeled precursor to an unexpected product. In addition to their role in mapping metabolic fates, isotopic labels also provide critical insight into pathway dynamics (i.e., metabolic fluxes) that cannot be obtained from conventional label-free metabolomic analyses. When labeling is compared quantitatively between conditions, for example, isotopic tracers can enable relative pathway activities to be inferred. To discover unexpected chemical transformations or unanticipated differences in metabolic pathway activities, we have developed X13CMS, a platform for analyzing liquid chromatography/mass spectrometry (LC/MS) data at the systems level. After providing cells, animals, or patients with an isotopically enriched metabolite (e.g., 13C, 15N, or 2H), X13CMS identifies compounds that have incorporated the isotopic tracer and reports the extent of labeling for each. The analysis can be performed with a single condition, or isotopic fates can be compared between multiple conditions. The choice of which metabolite to enrich and which isotopic label to use is highly context dependent, but 13C-glucose and 13C-glutamine are often applied because they feed a large number of metabolic pathways. X13CMS is freely available.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
A zip file with example data can be downloaded from http://pattilab.wustl.edu/software/x13cms/x13cms.php or http://pattilab.wustl.edu/download/Example.zip.
Code availability
X13CMS is freely available at http://pattilab.wustl.edu/software/x13cms/x13cms.php.
References
Guggenheim, K. Y. Rudolf Schoenheimer and the concept of the dynamic state of body constituents. J. Nutr. 121, 1701–1704 (1991).
Bloch, K. & Rittenberg, D. On the utilization of acetic acid for cholesterol formation. J. Biol. Chem. 145, 625–636 (1942).
Chance, E. M., Seeholzer, S. H., Kobayashi, K. & Williamson, J. R. Mathematical analysis of isotope labeling in the citric acid cycle with applications to 13C NMR studies in perfused rat hearts. J. Biol. Chem. 258, 13785–13794 (1983).
Malloy, C. R., Sherry, A. D. & Jeffrey, F. M. Carbon flux through citric acid cycle pathways in perfused heart by 13C NMR spectroscopy. FEBS Lett. 212, 58–62 (1987).
Plentl, A. & Schoenheimer, R. Studies in the metabolism of purines and pyrimidines by means of isotopic nitrogen. J. Biol. Chem. 153, 203–217 (1944).
Katz, J. & Wood, H. G. The use of glucose-C14 for the evaluation of the pathways of glucose metabolism. J. Biol. Chem. 235, 2165–2177 (1960).
Wilson, A. T. & Calvin, M. The photosynthetic cycle. CO 2 dependent transients. J. Am. Chem. Soc. 77, 5948–5957 (1955).
Calvin, M. & Benson, A. The path of carbon in photosynthesis. Science 107, 476–480 (1948).
Schoenheimer, R. & Rittenberg, D. The application of isotopes to the study of intermidiary metabolism. Science 87, 221–226 (1938).
Breitling, R., Pitt, A. R. & Barrett, M. P. Precision mapping of the metabolome. Trends Biotechnol. 24, 543–548 (2006).
Creek, D. J. et al. Stable isotope-assisted metabolomics for network-wide metabolic pathway elucidation. Anal. Chem. 84, 8442–8447 (2012).
Fan, T. W. M. et al. Stable isotope-resolved metabolomics and applications for drug development. Pharmacol. Ther. 133, 366–391 (2012).
Wilkinson, D. J. Historical and contemporary stable isotope tracer approaches to studying mammalian protein metabolism. Mass Spectrom. Rev. 37, 57–80 (2018).
Weindl, D., Wegner, A. & Hiller, K. Metabolome-wide analysis of stable isotope labeling-is it worth the effort? Front. Physiol. 6, 344 (2015).
Zamboni, N., Fendt, S.-M., Rühl, M. & Sauer, U. 13C-based metabolic flux analysis. Nat. Protoc. 4, 878–892 (2009).
Yuan, J., Bennett, B. D. & Rabinowitz, J. D. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat. Protoc. 3, 1328–1340 (2008).
Patti, G. J., Yanes, O. & Siuzdak, G. Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263–269 (2012).
da Silva, R. R., Dorrestein, P. C. & Quinn, R. A. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. USA 112, 12549–12550 (2015).
Dunn, W. B. et al. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 9, 44–66 (2013).
Mahieu, N. G. & Patti, G. J. Systems-level annotation of a metabolomics data set reduces 25000 features to fewer than 1000 unique metabolites. Anal. Chem. 89, 10397–10406 (2017).
Dias, D. A. et al. Current and future perspectives on the structural identification of small molecules in biological systems. Metabolites 6, (2016).
Kind, T. & Fiehn, O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2, 23–60 (2010).
Bingol, K. & Brüschweiler, R. Knowns and unknowns in metabolomics identified by multidimensional NMR and hybrid MS/NMR methods. Curr. Opin. Biotechnol. 43, 17–24 (2017).
Huang, X. et al. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal. Chem. 86, 1632–1639 (2014).
Zamboni, N., Saghatelian, A. & Patti, G. J. Defining the metabolome: size, flux, and regulation. Mol. Cell 58, 699–706 (2015).
Milne, S. B., Mathews, T. P., Myers, D. S., Ivanova, P. T. & Brown, H. A. Sum of the parts: mass spectrometry-based metabolomics. Biochemistry 52, 3829–3840 (2013).
Zhu, Z.-J. et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 8, 451–460 (2013).
Patti, G. J. Separation strategies for untargeted metabolomics. J. Sep. Sci. 34, 3460–3469 (2011).
Buescher, J. M. et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 34, 189–201 (2015).
Mahieu, N. G., Genenbacher, J. L. & Patti, G. J. A roadmap for the XCMS family of software solutions in metabolomics. Curr. Opin. Chem. Biol. 30, 87–93 (2016).
Tautenhahn, R., Patti, G. J., Rinehart, D. & Siuzdak, G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039 (2012).
Forsberg, E. M. et al. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat. Protoc. 13, 633–651 (2018).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Tautenhahn, R., Böttcher, C. & Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9, 504 (2008).
Prince, J. T. & Marcotte, E. M. Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Anal. Chem. 78, 6140–6152 (2006).
Mackie, A., Keseler, I. M., Nolan, L., Karp, P. D. & Paulsen, I. T. Dead end metabolites—defining the known unknowns of the E. coli metabolic network. PLoS ONE 8, e75210 (2013).
Chen, Y.-J. et al. Lactate metabolism is associated with mammalian mitochondria. Nat. Chem. Biol. 12, 937–943 (2016).
Faubert, B. et al. Lactate metabolism in human lung tumors. Cell 171, 358–371.e9 (2017).
Hui, S. et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017).
Gelman, S. J. et al. Evidence that 2-hydroxyglutarate is not readily metabolized in colorectal carcinoma cells. Cancer Metab. 3, 13 (2015).
Büscher, J. M., Czernik, D., Ewald, J. C., Sauer, U. & Zamboni, N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal. Chem. 81, 2135–2143 (2009).
Villas-Bôas, S. G., Mas, S., Åkesson, M., Smedsgaard, J. & Nielsen, J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 24, 613–646 (2005).
Roberts, L. D., Souza, A. L., Gerszten, R. E. & Clish, C. B. Targeted metabolomics. Curr. Protoc. Mol. Biol. Chapter 30, Unit 30.2.1–24 (2012).
Yao, C.-H. et al. Exogenous fatty acids are the preferred source of membrane lipids in proliferating fibroblasts. Cell Chem. Biol. 23, 483–493 (2016).
Hiller, K., Metallo, C. M., Kelleher, J. K. & Stephanopoulos, G. Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal. Chem. 82, 6621–6628 (2010).
Weindl, D., Wegner, A. & Hiller, K. MIA: non-targeted mass isotopolome analysis. Bioinformatics 32, 2875–2876 (2016).
Hoffmann, F., Jaeger, C., Bhattacharya, A., Schmitt, C. A. & Lisec, J. Nontargeted identification of tracer incorporation in high-resolution mass spectrometry. Anal. Chem. 90, 7253–7260 (2018).
Kiefer, P. et al. DynaMet: a fully automated pipeline for dynamic LC–MS data. Anal. Chem. 87, 9679–9686 (2015).
Kiefer, P., Schmitt, U. & Vorholt, J. A. eMZed: an open source framework in Python for rapid and interactive development of LC/MS data analysis workflows. Bioinformatics 29, 963–964 (2013).
Chokkathukalam, A. et al. mzMatch-ISO: an R tool for the annotation and relative quantification of isotope-labelled mass spectrometry data. Bioinformatics 29, 281–283 (2013).
Capellades, J. et al. geoRge: a computational tool to detect the presence of stable isotope labeling in LC/MS-based untargeted metabolomics. Anal. Chem. 88, 621–628 (2016).
Scheltema, R. A., Jankevics, A., Jansen, R. C., Swertz, M. A. & Breitling, R. PeakML/mzMatch: a file format, java library, r library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 83, 2786–2793 (2011).
Mahieu, N. G., Huang, X., Chen, Y.-J. & Patti, G. J. Credentialing features: a platform to benchmark and optimize untargeted metabolomic methods. Anal. Chem. 86, 9583–9589 (2014).
Bueschl, C. et al. MetExtract: a new software tool for the automated comprehensive extraction of metabolite-derived LC/MS signals in metabolomics research. Bioinformatics 28, 736–738 (2012).
Leeming, M. G. et al. High-resolution twin-ion metabolite extraction (HiTIME) mass spectrometry: nontargeted detection of unknown drug metabolites by isotope labeling, liquid chromatography mass spectrometry, and automated high-performance computing. Anal. Chem. 87, 4104–4109 (2015).
Su, X., Lu, W. & Rabinowitz, J. D. Metabolite spectral accuracy on Orbitraps. Anal. Chem. 89, 5940–5948 (2017).
Gelman, S. J. & Patti, G. J. Profiling cancer metabolism at the ‘omic’ level: a last resort or the next frontier?. Cancer Metab. 4, 2 (2016).
Cotter, D. G. et al. Ketogenesis prevents diet-induced fatty liver injury and hyperglycemia. J. Clin. Invest. 124, 5175–5190 (2014).
Yao, C.-H. et al. Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation. PLoS Biol. 16, e2003782 (2018).
Rashmi, R. et al. Radioresistant cervical cancers are sensitive to inhibition of glycolysis and redox metabolism. Cancer Res. 78, 1392–1403 (2018).
d’Avignon, D. A. et al. Hepatic ketogenic insufficiency reprograms hepatic glycogen metabolism and the lipidome. JCI Insight 3, 99762 (2018).
Kurczy, M. E. et al. Global isotope metabolomics reveals adaptive strategies for nitrogen assimilation. ACS Chem. Biol. 11, 1677–1685 (2016).
Johnson, C. H. et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21, 891–897 (2015).
Mahieu, N. G., Spalding, J. L., Gelman, S. J. & Patti, G. J. Defining and detecting complex peak relationships in mass spectral data: the Mz.unity algorithm. Anal. Chem. 88, 9037–9046 (2016).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Patti, G. J., Tautenhahn, R. & Siuzdak, G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 7, 508–516 (2012).
Libiseller, G. et al. IPO: a tool for automated optimization of XCMS parameters. BMC Bioinformatics 16, 118 (2015).
Cho, K. et al. isoMETLIN: a database for isotope-based metabolomics. Anal. Chem. 86, 9358–9361 (2014).
Millard, P., Letisse, F., Sokol, S. & Portais, J.-C. IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics 28, 1294–1296 (2012).
Acknowledgements
G.J.P. received financial support for this work from National Institutes of Health grants R35ES028365, U01CA235482, and R24OD024624, as well as the Alfred P. Sloan Foundation, the Pew Scholars Program in the Biomedical Sciences, and the Edward Mallinckrodt, Jr., Foundation. We thank N.G. Mahieu for his contributions to Figure 2.
Author information
Authors and Affiliations
Contributions
All authors equally developed and tested the protocol based on ref. 24. All authors drafted the manuscript and contributed to manuscript editing and revision.
Corresponding author
Ethics declarations
Competing interests
G.J.P. is a scientific advisory board member for Cambridge Isotope Laboratories and a recipient of the 2017 Agilent Early Career Professor Award. The remaining authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Huang, X. et al. Anal. Chem. 86, 1632–1639 (2014): https://pubs.acs.org/doi/10.1021/ac403384n
Key data used in this protocol
A zip file with example data can be downloaded from http://pattilab.wustl.edu/software/x13cms/x13cms.php or http://pattilab.wustl.edu/download/Example.zip
Supplementary information
Supplementary Methods
Representative example of preparing labeled samples for untargeted metabolomics.
Rights and permissions
About this article
Cite this article
Llufrio, E.M., Cho, K. & Patti, G.J. Systems-level analysis of isotopic labeling in untargeted metabolomic data by X13CMS. Nat Protoc 14, 1970–1990 (2019). https://doi.org/10.1038/s41596-019-0167-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0167-1
This article is cited by
-
A guide to precise measurements of isotope abundance by ESI-Orbitrap MS
Nature Protocols (2024)
-
Small molecule metabolites: discovery of biomarkers and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
Using mass spectrometry imaging to map fluxes quantitatively in the tumor ecosystem
Nature Communications (2023)
-
Global stable-isotope tracing metabolomics reveals system-wide metabolic alternations in aging Drosophila
Nature Communications (2022)
-
A guide to interrogating immunometabolism
Nature Reviews Immunology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.