Abstract
The 5-hydroxymethylcytosine binding, embryonic stem-cell-specific (HMCES) protein forms a covalent DNA-protein cross-link (DPC) with abasic (AP) sites in single-stranded DNA, and the resulting HMCES-DPC is thought to suppress double-strand break formation in S phase. However, the dynamics of HMCES cross-linking and whether any DNA repair pathways normally include an HMCES-DPC intermediate remain unknown. Here, we use Xenopus egg extracts to show that an HMCES-DPC forms on the AP site generated during replication-coupled DNA interstrand cross-link repair. We show that HMCES cross-links form on DNA after the replicative CDC45-MCM2-7-GINS (CMG) helicase has passed over the AP site, and that HMCES is subsequently removed by the SPRTN protease. The HMCES-DPC suppresses double-strand break formation, slows translesion synthesis past the AP site and introduces a bias for insertion of deoxyguanosine opposite the AP site. These data demonstrate that HMCES-DPCs form as intermediates in replication-coupled repair, and they suggest a general model of how HMCES protects AP sites during DNA replication.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the article and its Supplementary Information files. Sequencing read counts are available in Supplementary Tables 2–5. Source data showing unprocessed and uncropped gel and blot images are provided with this paper.
References
Krokan, H. E. & Bjørås, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 5, a012583 (2013).
Lindahl, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993).
Mohni, K. N. et al. HMCES maintains genome integrity by shielding abasic sites in single-strand DNA. Cell 176, 144–153.e13 (2019).
Thompson, P. S., Amidon, K. M., Mohni, K. N., Cortez, D. & Eichman, B. F. Protection of abasic sites during DNA replication by a stable thiazolidine protein-DNA cross-link. Nat. Struct. Mol. Biol. 26, 613–618 (2019).
Aravind, L., Anand, S. & Iyer, L. M. Novel autoproteolytic and DNA-damage sensing components in the bacterial SOS response and oxidized methylcytosine-induced eukaryotic DNA demethylation systems. Biol. Direct 8, 20 (2013).
Halabelian, L. et al. Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition. Nat. Struct. Mol. Biol. 26, 607–612 (2019).
Wang, N. et al. Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK. Nucleic Acids Res. 47, 10388–10399 (2019).
Srivastava, M. et al. HMCES safeguards replication from oxidative stress and ensures error-free repair. EMBO Rep. 21, e49123 (2020).
Mehta, K. P. M., Lovejoy, C. A., Zhao, R., Heintzman, D. R. & Cortez, D. HMCES maintains replication fork progression and prevents double-strand breaks in response to APOBEC deamination and abasic site formation. Cell Rep. 31, 107705 (2020).
Biayna, J. et al. Loss of the abasic site sensor HMCES is synthetic lethal with the activity of the APOBEC3A cytosine deaminase in cancer cells. PLoS Biol. 19, e3001176 (2021).
Price, N. E. et al. Interstrand DNA-DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J. Am. Chem. Soc. 136, 3483–3490 (2014).
Semlow, D. R. & Walter, J. C. Mechanisms of vertebrate DNA interstrand cross-link repair. Annu. Rev. Biochem. https://doi.org/10.1146/annurev-biochem-080320-112510 (2021).
Semlow, D. R., Zhang, J., Budzowska, M., Drohat, A. C. & Walter, J. C. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167, 498–511.e14 (2016).
Wu, R. A. et al. TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 567, 267–272 (2019).
Fullbright, G., Rycenga, H. B., Gruber, J. D. & Long, D. T. p97 Promotes a conserved mechanism of helicase unloading during DNA cross-link repair. Mol. Cell. Biol. 36, 2983–2994 (2016).
Knipscheer, P. et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 (2009).
Long, D. T., Räschle, M., Joukov, V. & Walter, J. C. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333, 84–87 (2011).
Räschle, M. et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 (2008).
Li, N. et al. Cooperation of the NEIL3 and Fanconi anemia/BRCA pathways in interstrand crosslink repair. Nucleic Acids Res. 48, 3014–3028 (2020).
Duxin, J. P., Dewar, J. M., Yardimci, H. & Walter, J. C. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159, 346–357 (2014).
Cimprich, K. A. & Cortez, D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 (2008).
Sparks, J. L. et al. The CMG helicase bypasses DNA-protein cross-links to facilitate their repair. Cell 176, 167–181.e21 (2019).
Rycenga, H. B., Wolfe, K. B., Yeh, E. S. & Long, D. T. Uncoupling of p97 ATPase activity has a dominant negative effect on protein extraction. Sci. Rep. 9, 10329 (2019).
Conti, B. A. & Smogorzewska, A. Mechanisms of direct replication restart at stressed replisomes. DNA Repair 95, 102947 (2020).
Larsen, N. B. et al. Replication-coupled DNA-protein crosslink repair by SPRTN and the proteasome in Xenopus egg extracts. Mol. Cell 73, 574–588.e7 (2019).
Kim, M. S. et al. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic Acids Res. 41, 1661–1668 (2013).
Reinking, H. K. et al. DNA structure-specific cleavage of DNA-protein crosslinks by the SPRTN protease. Mol. Cell 80, 102–113.e6 (2020).
Long, D. T., Joukov, V., Budzowska, M. & Walter, J. C. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol. Cell 56, 174–185 (2014).
Li, M. & Wilson, D. M. Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal 20, 678–707 (2014).
Marenstein, D. R., Wilson, D. M. & Teebor, G. W. Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA Repair 3, 527–533 (2004).
Rosenbaum, J. C. et al. The Rad51 paralogs facilitate a novel DNA strand specific damage tolerance pathway. Nat. Commun. 10, 3515 (2019).
Stingele, J. et al. Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol. Cell 64, 688–703 (2016).
Serbyn, N. et al. The aspartic protease Ddi1 contributes to DNA-protein crosslink repair in yeast. Mol. Cell 77, 1066–1079.e9 (2020).
Yip, M. C. J., Bodnar, N. O. & Rapoport, T. A. Ddi1 is a ubiquitin-dependent protease. Proc. Natl Acad. Sci. USA 117, 7776–7781 (2020).
Gallina, I. et al. The ubiquitin ligase RFWD3 is required for translesion DNA synthesis. Mol. Cell 81, 442–458.e9 (2021).
Enoiu, M., Ho, T. V., Long, D. T., Walter, J. C. & Schärer, O. D. Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol. Biol. 920, 203–219 (2012).
Zhang, J. et al. DNA interstrand cross-link repair requires replication-fork convergence. Nat. Struct. Mol. Biol. 22, 242–247 (2015).
Sparks, J. & Walter, J. C. Extracts for analysis of DNA replication in a nucleus-free system. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot097154 (2019).
Lebofsky, R., Takahashi, T. & Walter, J. C. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol. Biol. 521, 229–252 (2009).
Dewar, J. M., Budzowska, M. & Walter, J. C. The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350 (2015).
Budzowska, M., Graham, T. G., Sobeck, A., Waga, S. & Walter, J. C. Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link. EMBO J. 34, 1971–1985 (2015).
Acknowledgements
We thank J. Campbell, W. Dunphy and members of the Semlow and Walter laboratories for comments on the manuscript. D.R.S. is supported by NIH grant no. GM129422. J.C.W. is supported by NIH grant no. HL098316 and gift from the family of J.G. Wiseman. D.R.S. is a Ronald and JoAnne Willens Scholar. J.C.W. is a Howard Hughes Medical Institute Investigator and an American Cancer Society Research Professor.
Ethics declarations
Competing interests
D.R.S. and V.A.M. declare no competing interests. J.C.W. is a cofounder of MOMA Therapeutics, in which he has a financial interest.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Orlando Schärer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 An HMCES-DPC shields AP sites.
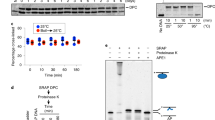
a, Purified recombinant FLAG-tagged Xenopus laevis HMCES proteins were resolved by SDS-PAGE and visualized by staining with InstantBlue. rHMCES∆PIP harbors W321A and L322A mutations that disrupt a conserved PIP-box that was previously show to mediate interaction with PCNA3. Asterisk, contaminating bands. b, A 5’ end radiolabeled 20mer oligonucleotide with a single deoxyuracil (-AP site) or AP site (+AP site) was incubated with rHMCES proteins shown in (a) for 60 min. Samples were then resolved on a denaturing polyacrylamide gel and visualized by autoradiography. Reactions contained 1 nM oligonucleotide and 50 nM rHMCES. c, Schematic of species produced by digestion of pICLAP replication intermediates with HincII and APE1. Digestion with HincII generates a 5.6 kb linear plasmid species while additional AP site cleavage by APE1 is expected to generate 2.3 kb and 3.3 kb species. d, HMCES immunodepletion. The extracts used in the reactions shown in e were blotted for HMCES. Asterisk, nonspecific band. e, pICLAP was replicated with [α-32P]dATP in the indicated egg extracts (shown in d). Samples were treated with proteinase K, phenol:chloroform extracted, and digested with HincII or with HincII and APE1. Digested DNAs were resolved on a native agarose gel and visualized by autoradiography. X structures indicate HincII-digested plasmids before ICL unhooking. f, Quantification of APE1 cleavage efficiency for the reactions shown in e. Cleavage efficiency was quantified as the intensity (Int) of 2.3 kb and 3.3 kb fragment bands in each lane divided by the total intensity of linear species bands ([Int2.3kB + Int3.3kB]/[Int2.3kB + Int3.3kB + Int5.6kB]). The efficiency of HMCES-DPC formation in mock-depleted extract was estimated by subtracting the extent of rAPE1 cleavage in mock-depleted extract from the extent of cleavage in HMCES-depleted extract at 45 min (arrowheads), when the absolute signal resulting from rAPE1 cleavage is maximal.
Extended Data Fig. 2 HMCES suppresses DSB formation specifically during NEIL3-dependent ICL repair.
a, b, d, f, and h, HMCES and NEIL3 immunodepletions. The extracts used in Fig. 2a (a), Extended Data Fig. 2c (b), Extended Data Fig. 2e (d), Extended Data Fig. 2g (f), and Fig. 2c (h) were blotted for HMCES and NEIL3 as indicated. Asterisks, non-specific bands. c, pICLAP was replicated with [α-32P]dATP in mock- or HMCES-depleted extracts. Replication intermediates were analyzed as in Fig. 2a alongside pCtrl replication products that were linearized by digestion with HincII. e, pICLAP was replicated with [α-32P]dATP in mock- or HMCES-depleted extracts supplemented with rHMCES, as indicated. Replication intermediates were analyzed as in Fig. 2a. g, pCtrl, pICLPt, or pICLAP were replicated with [α-32P]dATP in mock- or HMCES-depleted extract and replication intermediates were analyzed as in Fig. 2a.
Extended Data Fig. 3 HMCES suppresses ATR-dependent CHK1 phosphorylation during NEIL3-dependent ICL repair.
a, b, and d, HMCES immunodepletions. The extracts used in Fig. 2d (a), Extended Data Fig. 3c (b), and Extended Data Fig. 3e (d) were blotted for HMCES. Asterisks, non-specific bands. c, pICLAP was replicated in mock- or HMCES-depleted extracts supplemented with ATR inhibitor AZD6738 (ATRi), as indicated. Replication reactions were separated by SDS-PAGE and blotted for phospho-CHK1 and MCM6 (loading control). Accumulation of phosphorylated CHK1 was blocked by AZD6738 treatment, indicating that CHK1 phosphorylation is dependent on ATR. e, pCtrl, pICLPt, or pICLAP was replicated in mock- or HMCES-depleted extracts, as indicated. Replication reactions were analyzed as in c. HMCES depletion increased the accumulation of phosphorylated CHK1 only during replication of pICLAP, indicating a specific role for HMCES in NEIL3-dependent ICL repair.
Extended Data Fig. 4 TLS inhibition does not enhance DSB formation during NEIL3-dependent ICL repair.
a, REV1 and HMCES immunodepletion. The extracts used in the replication reactions shown in b were blotted for REV1 and HMCES. Asterisks, non-specific bands. b, pICLAP was replicated with [α-32P]dATP in the indicated extracts (shown in a). Replication intermediates were analyzed as in Fig. 2a.
Extended Data Fig. 5 Synchronization of ICL unhooking by NEIL3-depletion and add back.
a, NEIL3 and HMCES immunodepletion. The extracts used in the replication reactions shown in Fig. 3b were blotted for HMCES and NEIL3. Asterisks, non-specific bands. b, pICLAP was replicated in the presence of [α-32P]dATP in the indicated extracts (shown in a) supplemented with p97i for 60 min. rNEIL3 was then added to the reactions to allow ICL unhooking. Replication intermediates were analyzed as in Fig. 2a. c, HMCES immunodepletion. The extracts used in Fig. 4 were blotted for HMCES. Asterisk, non-specific band.
Extended Data Fig. 6 Nucleotide identity opposite the AP site does not effect NEIL3-dependent ICL unhooking or TLS.
a, Model of two alternative mechanisms of AP site bypass. Translesion synthesis (left branch) uses a specialized TLS polymerase for untemplated insertion of a nucleotide opposite the non-coding AP site. In this pathway, the nucleotide opposite the AP site in the parental plasmid does not influence insertion by the TLS polymerase. Template switching (right branch) uses the newly synthesized DNA of the other sister chromatid as a template for error-free bypass of the AP site. In this case, the inserted nucleotide is expected to co-vary with the nucleotide opposite the AP site in the parental plasmid. b, Plasmid design for next generation sequencing. We prepared four different plasmids, each containing an AP-ICL and a different nucleotide positioned opposite the AP site (Y), as well as a unique barcode (X-Y). An Nt.BstNBI restriction site allows specific cleavage of the AP site-containing strand to prevent its amplification during PCR. A dC-dC mismatch allows reads produced from amplification of the nascent strand that has bypassed the AP site (PCR product in orange box) to be distinguished from reads derived from the corresponding parental strand (upper PCR product). c, The four AP-ICL plasmids, each containing a different nucleotide opposite the AP-site (described in b), were replicated in undepleted egg extract supplemented with [α-32P]dATP. Replication intermediates were analyzed as in Fig. 2a. The base opposite the AP site had no apparent effect on replication or repair efficiency. d, HMCES immunodepletion. The extracts used to generate the sequencing libraries described in Fig. 5 were blotted for HMCES. Asterisk, non-specific band. e, The four AP-ICL plasmids described in b were pooled and replicated with [α-32P]dATP in the same extracts (shown in panel d) used to generate sequencing libraries described in Fig. 5. Replication intermediates were analyzed as in Fig. 2a. f, Analysis of PCR amplicons used for sequencing. In parallel to the reactions shown in e, pooled pICLAP plasmids were replicated in the indicated extracts (shown in d, but lacking [α-32P]dATP) supplemented with rHMCES, as indicated. DNA was extracted and digested with Nt.BstNBI (to cleave AP site-containing strands). The region of the replicated plasmids surrounding the ICL was then amplified by PCR. PCR amplicons were resolved by native agarose gel electrophoresis and visualized by Sybr Gold staining. g, Analysis of sequencing reads derived from individual pICLAP plasmids. The barcode was used to distinguish sequencing reads derived from the four different AP-ICL containing plasmids described in b. For each extract condition, we obtained >30,000 mapped reads, the vast majority (87.4%-87.8%) of which either perfectly matched the reference sequence or had a single point mutation corresponding to the position opposite the AP site. Of these reads, >11,000 in each condition derived from the nascent DNA stand produced upon bypass of the AP site. The fraction of reads corresponding to insertion of a given nucleotide opposite the AP site are plotted for each plasmid and extract condition. n, number of pooled nascent strand reads obtained for each condition. The result shows that the nucleotide opposite the AP site in the parental plasmid template does not influence the distribution of nascent DNA nucleotides inserted opposite the AP site after unhooking. Next generation sequencing read counts can be found in Supplementary Tables 2–5.
Extended Data Fig. 7 SPRTN protease activity is required for HMCES removal.
a and c, SPRTN immunodepletions. The extracts used in the replication reactions shown in Extended Data Fig. 7b (a) and Extended Data Fig. 7d (c) were blotted for SPRTN. b, pICLAP was replicated in mock- or SPRTN-depleted egg extract supplemented with wild-type (WT) or catalytically defective E89Q-mutated (EQ) rSPRTN, as indicated. Chromatin was recovered under stringent conditions, treated with the deubiquitylating enzyme USP21, and associated proteins were separated by SDS-PAGE and blotted for HMCES. Asterisk, non-specific band. d, pCtrl, pICLPt, or pICLAP were replicated in SPRTN-depleted egg extract supplemented with proteasome inhibitor MG262. Chromatin was analyzed as in Fig. 6c. Chromatin-associated HMCES is only observed in the pICLAP replication reaction, implying a specific role for HMCES in NEIL3-dependent ICL repair. Asterisk, non-specific band.
Extended Data Fig. 8 SPRTN-dependent proteolysis of the HMCES-DPC is not required for TLS past the AP site.
a, SPRTN immunodepletion. The extracts used in b-f were blotted for SPRTN. b, A plasmid containing a methylated HpaII-DPC that is refractory to degradation by the proteasome (pDPCme) or pICLAP were replicated with [α-32P]dATP in mock- or SPRTN-depleted egg extracts supplemented with p97i. Replication intermediates were analyzed as in Fig. 4a. c, Left, schematic of nascent strands generated during DPC repair. FspI and AatII cut 70 nucleotides to the left and 197 nucleotides to the right of the ICL, respectively, generating characteristic -30 stall, -1 to +1 stall, and strand extension products. Right, pDPCme was replicated as in b and nascent DNA strands were isolated, digested with FspI and AatII, and resolved by denaturing polyacrylamide gel electrophoresis. As previously reported, SPRTN-depletion delays TLS past the methylated HpaII-DPC, as evidenced by the persistence of rightward fork -1, 0, and +1 stall products and a delay in formation of rightward fork extension products. d, The persistence of the rightward fork -1, 0, and +1 stall products in c was quantified by dividing the summed intensity of the -1, 0, and +1 stall product bands in each lane by the intensity of the full-length rightward extension product band. Quantifications were normalized to the accumulation of the -1, 0, and +1 stall products at the 30 min time timepoint. Quantifications from two independent experiments are shown. e, Left, schematic of nascent strands generated during AP-ICL repair, as in Fig. 4b. Right, pICLAP was replicated as in b and nascent DNA strands were analyzed as in Fig. 4b. In contrast to TLS past the methylated HpaII-DPC shown in c, SPRTN-depletion accelerated TLS past the HMCES-DPC, as evidenced by the faster disappearance of rightward leading strand -1 stall products. This result indicates that TLS past the AP site does not require SPRTN-dependent proteolysis of the HMCES-DPC formed during NEIL3-dependent ICL repair. f, The persistence of the rightward fork -1 stall product in e was quantified as in d. Quantifications from two independent experiments are shown.
Extended Data Fig. 9 A proposed general model for AP site protection by HMCES during DNA replication.
(i) AP sites encountered in the leading strand template due to spontaneous depurination/depyrimidination or incomplete BER are bypassed by CMG. This leads to uncoupling of DNA unwinding by CMG from leading strand DNA synthesis by Pol ε and HMCES-DPC formation at a ssDNA/dsDNA junction. (ii) AP sites in the lagging strand template are bypassed without CMG uncoupling, and HMCES cross-links to the AP site fully embedded in ssDNA. In both scenarios, Pol δ then extends the lagging strand up to the HMCES-DPC, where upon synthesis stalls (iii). In both cases, the HMCES-DPC abuts a ssDNA/dsDNA junction, leading to proteolysis by SPRTN (iv). The HMCES-DPC and the peptide adduct generated after proteolysis stabilize the AP site until the lesion is bypassed by TLS (v) or an alternative, error-free mechanism (not depicted).
Supplementary information
Supplementary Tables
Supplementary Tables 1–5 in a single workbook.
Source data
Source Data Fig. 1
Unprocessed western blots and Southern blots presented in Fig. 1.
Source Data Fig. 2
Unprocessed western blots and gels presented in Fig. 2.
Source Data Fig. 3
Unprocessed gels presented in Fig. 3.
Source Data Fig. 4
Unprocessed gels presented in Fig. 4.
Source Data Fig. 6
Unprocessed western blots presented in Fig. 6.
Source Data Extended Data Fig. 1
Unprocessed western blots and gels presented in Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Unprocessed western blots and gels presented in Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Unprocessed western blots presented in Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Unprocessed western blots and gels presented in Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Unprocessed western blots and gels presented in Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Unprocessed western blots and gels presented in Extended Data Fig. 6.
Source Data Extended Data Fig. 7
Unprocessed western blots presented in Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Unprocessed western blots and gels presented in Extended Data Fig. 8.
Rights and permissions
About this article
Cite this article
Semlow, D.R., MacKrell, V.A. & Walter, J.C. The HMCES DNA-protein cross-link functions as an intermediate in DNA interstrand cross-link repair. Nat Struct Mol Biol 29, 451–462 (2022). https://doi.org/10.1038/s41594-022-00764-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00764-0
This article is cited by
-
DNA-Protein-Crosslinks: Schaden und Schutz zugleich
BIOspektrum (2023)