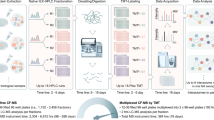
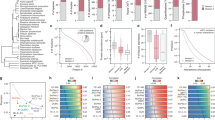
Abstract
Despite the availability of methods for analyzing protein complexes, systematic analysis of complexes under multiple conditions remains challenging. Approaches based on biochemical fractionation of intact, native complexes and correlation of protein profiles have shown promise. However, most approaches for interpreting cofractionation datasets to yield complex composition and rearrangements between samples depend considerably on protein–protein interaction inference. We introduce PCprophet, a toolkit built on size exclusion chromatography–sequential window acquisition of all theoretical mass spectrometry (SEC-SWATH-MS) data to predict protein complexes and characterize their changes across experimental conditions. We demonstrate improved performance of PCprophet over state-of-the-art approaches and introduce a Bayesian approach to analyze altered protein–protein interactions across conditions. We provide both command-line and graphical interfaces to support the application of PCprophet to any cofractionation MS dataset, independent of separation or quantitative liquid chromatography–MS workflow, for the detection and quantitative tracking of protein complexes and their physiological dynamics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The annotated training dataset (including the calculated features and manual annotation) for the core RF model of PCprophet, and the GO term files (AmiGO2 database; http://amigo.geneontology.org/amigo) are available at https://github.com/anfoss/PCprophet and on Zenodo: https://doi.org/10.5281/zenodo.4574600). The information of the original raw dataset for training PCprophet and the other two datasets used for benchmarking and differential analysis have been demonstrated in Supplementary Table 2. The protein interaction and complex data were downloaded directly from the STRING (https://string-db.org/cgi/download), CORUM (core complexes; available at http://mips.helmholtz-muenchen.de/corum/#download), BioGRID (https://downloads.thebiogrid.org/BioGRID) and BioPlex (https://bioplex.hms.harvard.edu/) databases. All protein structures referred in this study have been provided with their PDB (https://www.rcsb.org/) IDs (4GQB, 6J2C and 6J2Q). The proteomics DDA and PRM raw files are available in PRIDE59 (https://www.ebi.ac.uk/pride/) under the identifier PXD022175. Source Data for Figs. 2–4 are available with this paper. Source data for Supplementary Figs. 1–5 and 11 are available at https://github.com/anfoss/PCprophet. The data that support the findings of this study are also available from the corresponding author upon request.
Code availability
PCprophet is open-access and freely available for academic purposes at https://github.com/anfoss/PCprophet under the MIT License. For the packages used to analyze MS data in this study, please refer to the Supplementary Notes for more details.
References
Marsh, J. A. & Teichmann, S. A. Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 84, 551–575 (2015).
Pan, J. et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst. 6, 555–568 e557 (2018).
Sowmya, G., Breen, E. J. & Ranganathan, S. Linking structural features of protein complexes and biological function. Protein Sci. 24, 1486–1494 (2015).
Spirin, V. & Mirny, L. A. Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA 100, 12123–12128 (2003).
Salas, D., Stacey, R. G., Akinlaja, M. & Foster, L. J. Next-generation interactomics: considerations for the use of co-elution to measure protein interaction networks. Mol. Cell Proteom. 19, 1–10 (2020).
Crozier, T. W. M., Tinti, M., Larance, M., Lamond, A. I. & Ferguson, M. A. J. Prediction of protein complexes in Trypanosoma brucei by protein correlation profiling mass spectrometry and machine learning. Mol. Cell Proteom. 16, 2254–2267 (2017).
Heusel, M. et al. A global screen for assembly state changes of the mitotic proteome by SEC-SWATH-MS. Cell Syst. 10, 133–155.e6 (2019).
Hu, L. Z. et al. EPIC: software toolkit for elution profile-based inference of protein complexes. Nat. Methods 16, 737–742 (2019).
Kirkwood, K. J., Ahmad, Y., Larance, M. & Lamond, A. I. Characterization of native protein complexes and protein isoform variation using size-fractionation-based quantitative proteomics. Mol. Cell Proteom. 12, 3851–3873 (2013).
Scott, N. E. et al. Interactome disassembly during apoptosis occurs independent of caspase cleavage. Mol. Syst. Biol. 13, 906 (2017).
Heusel, M. et al. Complex-centric proteome profiling by SEC-SWATH-MS. Mol. Syst. Biol. 15, e8438 (2019).
McBride, Z. et al. A label-free mass spectrometry method to predict endogenous protein complex composition. Mol. Cell Proteom. 18, 1588–1606 (2019).
Stacey, R. G., Skinnider, M. A., Scott, N. E. & Foster, L. J. A rapid and accurate approach for prediction of interactomes from coelution data (PrInCE). BMC Bioinf. 18, 457 (2017).
Kerr, C. H. et al. Dynamic rewiring of the human interactome by interferon signaling. Genome Biol. 21, 140 (2020).
Pourhaghighi, R. et al. BraInMap elucidates the macromolecular connectivity landscape of mammalian brain. Cell Syst. 10, 333–350.e314 (2020).
Stacey, R. G., Skinnider, M. A. & Foster, L. J. On the robustness of graph-based clustering to random network alterations. Mol. Cell Proteom. 20, 100002 (2020).
Quinlan, R. C4.5: Programs for Machine Learning (Morgan Kaufmann, 1993).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Zhang, H. The optimality of naïve Bayes. in Proc. Seventeenth International Florida Artificial Intelligence Research Society Conference (AAAI Press, 2004).
Cortes, C. & Vapnik, V. Support-Vector Networks. Mach. Learn. 20, 273–297 (1995).
Lecessie, S. & Vanhouwelingen, J. C. Ridge estimators in logistic-regression. Appl Stat.-J. R. St C. 41, 191–201 (1992).
Giurgiu, M. et al. CORUM: the comprehensive resource of mammalian protein complexes – 2019. Nucleic Acids Res. 47, D559–D563 (2019).
Kristensen, A. R., Gsponer, J. & Foster, L. J. A high-throughput approach for measuring temporal changes in the interactome. Nat. Methods 9, 907–909 (2012).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Oughtred, R. et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019).
Havugimana, P. C. et al. A census of human soluble protein complexes. Cell 150, 1068–1081 (2012).
Livneh, I., Cohen-Kaplan, V., Cohen-Rosenzweig, C., Avni, N. & Ciechanover, A. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res 26, 869–885 (2016).
Lasker, K. et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl Acad. Sci. USA 109, 1380–1387 (2012).
Ding, Z. et al. Structural snapshots of 26S proteasome reveal tetraubiquitin-induced conformations. Mol. Cell 73, 1150–1161.e1156 (2019).
Huang, D. T. et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 33, 483–495 (2009).
Kohroki, J., Nishiyama, T., Nakamura, T. & Masuho, Y. ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett. 579, 6796–6802 (2005).
Lowe, N. et al. Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development 141, 3994–4005 (2014).
Collins, M. O. et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, 16–23 (2006).
Antonysamy, S. et al. Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl Acad. Sci. USA 109, 17960–17965 (2012).
Scoumanne, A., Zhang, J. & Chen, X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 37, 4965–4976 (2009).
Gu, Z. et al. The p44/wdr77-dependent cellular proliferation process during lung development is reactivated in lung cancer. Oncogene 32, 1888–1900 (2013).
Bludau, I. & Aebersold, R. Proteomic and interactomic insights into the molecular basis of cell functional diversity. Nat. Rev. Mol. Cell Biol. 21, 327–340 (2020).
Bludau, I. et al. Complex-centric proteome profiling by SEC-SWATH-MS for the parallel detection of hundreds of protein complexes. Nat. Protoc. 15, 2341–2386 (2020).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Rost, H. L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).
Rost, H. L. et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat. Methods 13, 777–783 (2016).
Dijkstra, E. W. A note on two problems in connexion with graphs. Numer. Math. 1, 3 (1959).
Vert, J. P, Tsuda, K & Schoelkopf, B. Kernel Methods in Computational Biology (MIT Press, 2004) 35–70.
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn Res 12, 2825–2830 (2011).
Frank, E., Hall, M. A., & Witten, I. H. The WEKA Workbench. Online Appendix for ‘Data Mining: Practical Machine Learning Tools and Techniques’, 4th edn (Morgan Kaufmann, 2016).
Matthews, B. W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta 405, 442–451 (1975).
Franz, M. et al. GeneMANIA update 2018. Nucleic Acids Res. 46, W60–W64 (2018).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Carbon, S. et al. AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 (2009).
The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Wang, J. Z., Du, Z., Payattakool, R., Yu, P. S. & Chen, C. F. A new method to measure the semantic similarity of GO terms. Bioinformatics 23, 1274–1281 (2007).
The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
McKinney, W. Data structure for statistical computation in Python. in The 9th Python in Science Conference (eds., Stéfan van der Walt and Jarrod Millman) 56–61 (2010).
Hagberg, A. A., Schult, D. A. & Swart, P. J. Exploring network structure, dynamics, and function using NetworkX. in The 7th Python in Science Conference (SciPy2008) (eds., Varoquaux, G. et al.) (2008).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Acknowledgements
This work was supported financially in part by the Swiss National Science Foundation (grant No. 3100A0-688 107679 to R.A.) and the European Research Council (ERC-20140AdG 670821 to R.A.). C.L. is currently supported by a National Health and Medicine Research Council (NHMRC) of Australia CJ Martin Early Career Research Fellowship (1143366). F.U., M.G. and F.F. were supported by the Innovative Medicines Initiative project ULTRA-DD (FP07/2007-2013, grant no. 115766). F.W. and B.W. were supported by the ETH strategic focus area ‘Personalized Health and Related Technologies (PHRT)’. M.G. acknowledges support from the EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN, grant no. 875510). I.B. acknowledges funding support from the Swiss National Science Foundation (31003A_166435). A.W.P. is supported by an NHMRC Principal Research Fellowship (1137739). We thank E. Milani for providing the vector pcDNA5/FRT/TO/SH/GW containing the ORF for GFP. We thank N. de Souza from ETH Zürich for her critical comments on this study.
Author information
Authors and Affiliations
Contributions
R.A., A.F., M. Heusel, C.L. and M.G. conceived and designed the project. A.F. and C.L. designed, developed and implemented PCprophet, and conducted data analysis, machine-learning prediction and benchmarking experiments with other existing methods. F.U. and F.W. performed experimental validation of the predicted complexes and interactions in the study. P.S. designed and implemented the differential analysis module of PCprophet for protein complexes. M. Heusel provided data and initial analysis. M. Heusel, F.F. and F.U. contributed to data annotation and provided critical feedback and comments on the biological aspects. M. Hallal contributed to EPIC performance comparison and reproducibility test of the PCprophet performance. I.B. assisted with benchmarking experiments with CCprofiler and provided useful insights. T.C., P.X., J.S., B.W. and A.W.P. provided critical and insightful comments during the development of PCprophet. C.L., A.F., M.G. and R.A. drafted the manuscript, which has been revised and approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks Stefan Tenzer and the other, anonymous, reviewers, for their contribution to the peer review of this work. Arunima Singh was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary Notes, Results, Figs. 1–11 and Tables 1–3.
Source data
Source Data Fig. 2
Source data for Fig 2.
Source Data Fig. 3
Source data for Fig 3.
Source Data Fig. 4
Source data for Fig 4.
Rights and permissions
About this article
Cite this article
Fossati, A., Li, C., Uliana, F. et al. PCprophet: a framework for protein complex prediction and differential analysis using proteomic data. Nat Methods 18, 520–527 (2021). https://doi.org/10.1038/s41592-021-01107-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01107-5
This article is cited by
-
DIP-MS: ultra-deep interaction proteomics for the deconvolution of protein complexes
Nature Methods (2024)
-
Tapioca: a platform for predicting de novo protein–protein interactions in dynamic contexts
Nature Methods (2024)
-
Next-generation proteomics for quantitative Jumbophage-bacteria interaction mapping
Nature Communications (2023)
-
Analysis of DIA proteomics data using MSFragger-DIA and FragPipe computational platform
Nature Communications (2023)
-
Improved in situ characterization of protein complex dynamics at scale with thermal proximity co-aggregation
Nature Communications (2023)