Abstract
N6-methyladenosine (m6A) is a widespread RNA modification that influences nearly every aspect of the messenger RNA lifecycle. Our understanding of m6A has been facilitated by the development of global m6A mapping methods, which use antibodies to immunoprecipitate methylated RNA. However, these methods have several limitations, including high input RNA requirements and cross-reactivity to other RNA modifications. Here, we present DART-seq (deamination adjacent to RNA modification targets), an antibody-free method for detecting m6A sites. In DART-seq, the cytidine deaminase APOBEC1 is fused to the m6A-binding YTH domain. APOBEC1-YTH expression in cells induces C-to-U deamination at sites adjacent to m6A residues, which are detected using standard RNA-seq. DART-seq identifies thousands of m6A sites in cells from as little as 10 ng of total RNA and can detect m6A accumulation in cells over time. Additionally, we use long-read DART-seq to gain insights into m6A distribution along the length of individual transcripts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study have been deposited in NCBI’s Gene Expression Omnibus under accession code GSE125780.
References
Zhao, B. S., Roundtree, I. A. & He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Meyer, K. D. & Jaffrey, S. R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Navaratnam, N. et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 268, 20709–20712 (1993).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Schibler, U., Kelley, D. E. & Perry, R. P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 115, 695–714 (1977).
Wei, C. M. & Moss, B. Nucleotide sequences at the N 6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676 (1977).
Wei, C. M., Gershowitz, A. & Moss, B. 5′-terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401 (1976).
Li, F., Zhao, D., Wu, J. & Shi, Y. Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res. 24, 1490–1492 (2014).
Wang, X. et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8, 284–296 (2014).
Chen, K. et al. High-resolution N 6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew. Chem. Int Ed. Engl. 54, 1587–1590 (2015).
Liu, N. et al. Probing N 6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Weng, Y. L. et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325 e316 (2018).
Merkurjev, D. et al. Synaptic N 6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014 (2018).
Zeng, Y. et al. Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 16, e2006092 (2018).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Luo, S. & Tong, L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl Acad. Sci. USA 111, 13834–13839 (2014).
Xu, C. et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 (2014).
Molinie, B. et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692–698 (2016).
Slobodin, B. et al. Transcription impacts the efficiency of mRNA translation via co-transcriptional N 6-adenosine methylation. Cell 169, 326–337 e312 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Shah, A., Qian, Y., Weyn-Vanhentenryck, S. M. & Zhang, C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 33, 566–567 (2017).
Wu, T. D. & Watanabe, C. K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Olarerin-George, A. O. & Jaffrey, S. R. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics 33, 1563–1564 (2017).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Lindenbaum, P. JVarkit: java-based utilities for bioinformatics. Figshare https://doi.org/10.6084/m9.figshare.1425030.v1 (2015).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
We thank S.H. Choi for generating METTL3-depleted cells. We also thank S. Horner and members of the Meyer laboratory for comments and suggestions. This study was supported by the National Institutes of Health grant nos. R00MH104712, R01MH118366 and DP1DA046584. K.D.M. is also supported by the Rita Allen Foundation, the Kinship Foundation Searle Scholars Program and the Klingenstein–Simons Fellowship in Neurosciences.
Author information
Authors and Affiliations
Contributions
K.D.M. conceived of the project, collected and analyzed the data, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Peer review information Nicole Rusk was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Synthesis of APOBEC1-YTH and APOBEC1-YTHmut for in vitro deaminase assays.
(a) Sanger sequencing chromatograms from in vitro deamination assays demonstrate that the cytidine adjacent to m6A is converted to uridine when m6A-containing RNA is used in the assay. APOBEC1-YTHmut, which lacks the m6A-binding portion of the YTH domain, fails to convert C to U in m6A-containing RNA. Chromatograms are representative images from 3 individual clones picked for sequencing after subjecting PCR products to TOPO cloning. (b) Western blotting for anti-HA was performed to indicate the levels of APOBEC1-YTH or APOBEC1-YTHmut in in vitro DART-Seq assays. APOBEC1-YTH or APOBEC1-YTHmut proteins were synthesized in vitro and an aliquot equivalent to the amount used for DART-Seq assays was removed and subjected to western blot analysis. General protein levels are also shown using ponceau S staining of the membrane. Images are representative of n = 2 independent experiments. (c) APOBEC1-YTHmut exhibits substantially reduced binding to m6A in RNA pulldown assays. Anti-HA western blot results are shown after mixing HA-tagged APOBEC1-YTH or APOBEC1-YTHmut with A- or m6A-containing biotinylated RNA and subsequently purifying RNA-bound protein with streptavidin pulldown. 2% of input is shown as a reference. Blots are representative of n = 2 independent experiments.
Supplementary Figure 2 Analysis of individual DART-Seq replicates.
Three individual DART-Seq replicates were analyzed for the number of C to U mutations per RNA. Pearson correlation coefficient indicated a high degree of overlap between individual replicates (r = 0.94 and above, as indicated on each plot; n = 3 independent samples).
Supplementary Figure 3 Characteristics of DART-Seq datasets.
(a) Metagene analysis showing a density plot of the absolute distance of C to U editing sites from the stop codon (position 0). APOBEC1-YTH expression leads to C to U editing enriched in the vicinity of the stop codon, which mirrors the distribution of m6A. In contrast, APOBEC1-YTHmut expression leads to editing throughout the 3’UTR (n = 3 independent samples per condition). (b) Pie chart showing the distribution of C to U editing sites in cells expressing APOBEC1 alone, which is characterized by a greater amount of intronic and intergenic editing events compared to APOBEC1-YTH-expressing cells (n = 2 independent samples). (c) Venn diagram showing overlapping C to U editing sites in DART-Seq libraries prepared from cells expressing APOBEC1-YTHmut or APOBEC1 alone. (d) Distribution of exon lengths for internal exons containing C to U editing sites in APOBEC1-YTH-expressing cells. Exon lengths of internal exons from the human genome (hg19 annotation) are shown for comparison. There is a greater proportion of long internal exons among C to U edited exons in APOBEC1-YTH-expressing cells compared to the natural distribution throughout the transcriptome.
Supplementary Figure 4 Examples of DART-Seq editing in cellular RNAs.
IGV browser tracks show read coverage in the SPEN (a) and SON (b) mRNAs, as well as the XIST (c) long non-coding RNA. C to U mutations found in at least 10% of reads are indicated by blue/red (C/U) coloring in the SPEN and SON transcripts (positive strand) and by gold/green (C/U) coloring in the XIST transcript (negative strand). C to U editing is shown for cells expressing APOBEC1-YTH (n = 3 independent samples), APOBEC1 alone (n = 2 independent samples), and APOBEC1-YTHmut (n = 3 independent samples). MeRIP-Seq tracks indicating m6A are shown in blue.
Supplementary Figure 5 Comparison of DART-Seq to miCLIP.
(a) DART-Seq sites were compared to m6A peak regions identified by immunoprecipitation-based m6A mapping from three different datasets (PA-m6A-Seq15, m6A-Seq4, and MeRIP-Seq3). In addition, CIMS/CITS sites from two single-nucleotide resolution miCLIP datasets5,6 were also compared to m6A peak regions from the three datasets. Shown is the overlap between DART-Seq and the two miCLIP datasets reported as the percentage of m6A peak sites that overlap. Across all three datasets (PA-m6A-Seq, m6A-Seq, and MeRIP-Seq), DART-Seq performs similarly to miCLIP in identifying m6A sites. (b) m6A sites not located within a DRACH consensus motif were taken from the Ke et al. 20156 dataset. DART-Seq C to U editing events which overlapped with CIMS/CITS sites at these non-DRACH m6A residues are shown as a percentage of the total number of non-DRACH m6A sites. CIMS/CITS sites from Linder et al. 20155 were also compared to these non-DRACH sites, showing a similar degree of overlap as is observed with DART-Seq. (c) Analysis of DART-Seq site clustering suggests minimal promiscuous C to U editing at non-m6A-adjacent cytidines. Individual DART-Seq or CIMS/CITS sites were analyzed from DART-Seq data or two miCLIP datasets (Ke et al. 2015 and Linder et al. 2015). For each site within the dataset, the closest C to U editing site (DART-Seq) or CIMS/CITS site (miCLIP) was identified. Shown is the proportion of total sites that are at least 10 nt or 20 nt away from the closest site. All three datasets show a similar distribution of sites, with the majority of sites being at least 10 nt apart.
Supplementary Figure 6 Comparison of DART-Seq to antibody-based m6A mapping.
(a) Frequency plot showing the distribution of DART-Seq C to U editing events relative to CIMS and CITS sites at cytidines adjacent to m6A from miCLIP data5 (left; ‘DART-Seq’). Each individual CIMS or CITS site from miCLIP was centered at 0, and the distance to the nearest C to U editing event from DART-Seq was identified and plotted according to its distance up/downstream of the miCLIP site. For comparison, the same distribution plot was generated using CIMS and CITS sites from a second independent miCLIP study6 (right; ‘m6A IP’). Both plots demonstrate that C to U editing sites (DART-Seq) or CIMS/CITS sites (m6A IP) are frequently enriched at the exact same cytidine that is marked by miCLIP. (b) Frequency plots as in (a) showing the distance of DART-Seq (left) or m6A-CLIP (right) sites within 20 nucleotides up/downstream of miCLIP CIMS/CITS sites. (c) Identity and frequency of the nucleotide immediately preceding C to U mutation sites in DART-Seq datasets from cells expressing APOBEC1-YTH or APOBEC1 alone. There is a strong preference for A residues preceding C to U editing sites in APOBEC1-YTH-expressing cells (n = 40,263 APOBEC1-YTH; n = 8,774 APOBEC1).
Supplementary Figure 7 Validation of DART-Seq editing adjacent to m6A sites.
(a) IGV browser tracks showing C to U editing in APOBEC1-YTH-expressing cells at m6A sites in the BSG mRNA (sites with >10% editing are indicated by blue/red (C/U) coloring at the site; BSG is on the positive strand). Editing is not present in cells expressing APOBEC1 alone. Expanded views show Sanger sequencing following RT-PCR from cells expressing APOBEC1-YTH or APOBEC1 alone. C to U editing events (*) near m6A sites identified by SCARLET16 are shown, along with m6A abundance at that site as measured by SCARLET. (b) IGV browser tracks show C to U editing in APOBEC1-YTH-expressing cells at m6A sites in the ACTB mRNA (sites with >10% editing are indicated by gold/green (C/U) coloring at the site; ACTB is transcribed from the negative strand). Editing is not present in APOBEC1 alone-expressing cells. Expanded views show Sanger sequencing following RT-PCR from cells expressing APOBEC1-YTH or APOBEC1 alone. C to U editing events (*) near m6A sites identified by miCLIP5 are shown. Results represent a minimum of n = 2 independent experiments.
Supplementary Figure 8 Validation of METTL3 depletion in HEK293T cells.
(a) m6A immunoblotting of total RNA from wild type (WT) and METTL3-depleted cells (knockdown; KD) confirms loss of m6A. (b) Western blotting of WT and KD cells shows loss of METTL3 in KD cells. (c) IGV browser view showing stranded RNA-seq coverage at the METTL3 locus in METTL3 KD cells. Reads corresponding to METTL3 (antisense strand) are shown in purple. The expanded view shows the region in exon 3 of METTL3 which is targeted by the sgRNA being expressed in METTL3 KD cells. Yellow highlighted region indicates the site of one or two base insertions (T or TT) in the majority of reads. These insertions are predicted to cause a frameshift and premature stop codon. Results represent a minimum of n = 3 independent experiments.
Supplementary Figure 9 DART-Seq in METTL3-depleted cells.
(a) Motif analysis was performed using regions +/− 4 nt surrounding C to U editing sites identified by DART-Seq in METTL3-depleted (METTL3 KD) cells. Shown is the most highly enriched motif. n = 8,112. P value was calculated using the cumulative binomial distribution. (b) Pie chart showing the distribution of C to U editing events identified by DART-Seq in METTL3 KD cells. n = 2 independent samples. (c) Venn diagram showing the overlap between C to U editing sites called by DART-Seq in wild type (WT) and METTL3 KD HEK293T cells. (d) IGV browser tracks showing read coverage and C to U mutations in wild type and METTL3 KD cells in two representative mRNAs (RPS4X and EEF2; both transcribed from the negative strand). C to U mutations found with at least 10% frequency are indicated by gold/green (C/U) coloring at the mutation site. MeRIP-Seq is shown in blue. C to U mutations in wild type cells are lost in METTL3 KD cells. Images represent 2 independent samples.
Supplementary Figure 10 Low-input DART-Seq.
(a) Venn diagrams showing overlap among methylated RNAs identified by DART-Seq from sequencing libraries prepared using 1μg, 100ng, and 10ng total RNA as input (n = 3, 1, and 1 independent samples, respectively). (b) IGV browser tracks showing read coverage and C to U mutations in three representative mRNAs (BSG, EEF2, and HSPA5). C to U mutations occurring with at least 10% frequency are indicated by blue/red (C/U) coloring for the BSG transcript (positive strand) and by gold/green (C/U) coloring for the EEF2 and HSPA5 transcripts (negative strand). Tracks shown are for DART-Seq experiments using 1μg (n =3), 100ng (n =1), or 10ng (n =1) total RNA as input from cells expressing APOBEC1-YTH. The APOBEC1 track indicates DART-Seq reads from cells expressing APOBEC1 alone and prepared using 1μg total RNA as input (n = 2). MeRIP-Seq track is shown in blue to indicate m6A identified by antibody-based methods. (c) Low-input DART-Seq performs comparably to high-input DART-Seq in its ability to identify m6A sites. DART-Seq datasets from samples prepared using 10ng (n =1), 100ng (n =1), and 1μg (n =3) of total RNA as input were compared to m6A peak regions from three independent m6A immunoprecipitation datasets (m6A-Seq4, PA-m6A-Seq15, and MeRIP-Seq3). The proportion of m6A peaks from each dataset that overlaps with each of the three DART-Seq datasets is shown. (d) Motif analysis showing the most highly enriched motifs surrounding C to U sites identified by DART-Seq using 10ng (n = 73,205) or 100ng (n = 67,310) of total RNA as input. P values for individual motif enrichment were calculated using the cumulative binomial distribution. (e) Metagene analysis of C to U editing sites from HEK293T cells expressing APOBEC1-YTH and using 10ng of total RNA for sequencing (n = 1 sequencing sample).
Supplementary Figure 11 Properties of APOBEC1-YTH expressing cells.
(a) APOBEC1-YTH expression does not affect cell viability. Trypan blue staining of HEK293T cells transfected with APOBEC1-YTH was used to assess cell viability. The proportion of viable cells is shown compared to untransfected HEK293T cells (“Control”). Shown are sample means +/- S.D. n = 4 biological replicates; “n.s” = non-significant (P = 0.943); 2-tailed t-test. (b) APOBEC1-YTH expression does not affect levels of the apoptosis marker cleaved caspase 3. HEK293T cells were transfected with APOBEC1-YTH and protein was isolated 24 h later. Western blotting was done to assess levels of cleaved caspase 3. Shown are the average levels of cleaved caspase 3 relative to β-actin. Compared to untransfected HEK293T cells (“Control”), APOBEC1-YTH-expressing cells do not exhibit altered levels of cleaved caspase 3. Shown are sample means +/- S.D. n = 4 biological replicates; “n.s” = non-significant (P = 0.462); 2-tailed t-test. (c) Levels of major m6A regulators are not altered by APOBEC1-YTH expression. Western blotting was used to assess levels of METTL3 and YTHDF2 in APOBEC1-YTH-transfected cells as well as untransfected cells (“Control”). Shown are representative images from n = 2 biological replicates. (d) APOBEC1-YTH exhibits cytoplasmic and weak nuclear expression in cells. HEK293T cells were transfected with APOBEC1-YTH, which contains a c-terminal HA tag. 24 h later, immunofluorescence was used to assess localization of APOBEC1-YTH using an anti-HA antibody. DAPI was used to mark cell nuclei. n = 2 independent experiments.
Supplementary Figure 12 In vitro DART-Seq identifies m6A sites in cellular RNA.
(a) IGV browser tracks show read coverage and C to U mutations in three representative mRNAs (JUN, HSPA5, and TMEM60; all transcribed from the negative strand). C to U mutations are indicated by gold/green (C/U) coloring, and total reads (RNA-Seq) are shown in gray. MeRIP-Seq peaks indicating regions of m6A are shown in blue. For each RNA, DART-Seq tracks from cells expressing APOBEC1-YTH or APOBEC1 alone (“In cells”; n = 3 and 2 independent samples, respectively) are shown next to DART-Seq tracks from in vitro deamination assays using HEK293T cell RNA (“In vitro”; n = 2 independent samples per condition).
Supplementary Figure 13 Properties of in vitro DART-Seq data.
(a) Metagene analysis showing a density plot of the absolute distance of C to U editing sites from the stop codon (position 0) for in vitro DART-Seq assays. C to U editing is enriched in the vicinity of the stop codon, which mirrors the distribution of m6A. n = 2 independent samples. (b) Motif analysis shows motifs found within a +/− 4 nt region surrounding C to U editing sites from in vitro DART-Seq datasets. The most enriched motif from in vitro DART-Seq assays using APOBEC1-YTH contains a GGACU sequence, which matches the m6A consensus (top; n = 9,428). In contrast, the top motif from in vitro DART-Seq assays using APOBEC1 alone fails to show enrichment for an m6A consensus motif (bottom; n = 12,206). P values were calculated using the cumulative binomial distribution.
Supplementary Figure 14 DART-Seq does not identify m6Am sites.
Regions surrounding DART-Seq sites overlap with a single predicted m6Am site in CDC42BPA. However, DART-Seq reads showing at least 10% C to U conversion (gold to green, respectively) indicate reduction of C to U editing in METTL3-depleted (METTL3 KD) cells, suggesting that this is an m6A and not an m6Am site. The putative m6A/m6Am site (yellow) is also located in a DRACH motif and internally within the 5’UTR. Percentages indicate proportion of C to U editing event at that site.
Supplementary Figure 15 DART-Seq estimates m6A abundance within RNAs.
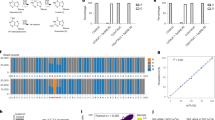
(a) In vitro deamination assays were performed using synthetic RNAs with various amounts of m6A (0%, 25%, 75%, or 100%). Sanger sequencing was used to determine the relative proportion of editing at the adjacent cytidine residue. n = 2 biological replicates. (b) Quantification of the proportion of U/C editing at the cytidine adjacent to the m6A site in (a) reveals a direct relationship between the amount of C to U editing and the abundance of m6A within the RNA.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15.
Supplementary Table 1: Sites identified by DART-seq.
Listed are the genome coordinates of DART-seq C-to-U editing sites identified from cells expressing APOBEC1 alone or APOBEC1-YTHmut. Also indicated are the number of C-to-U mutations at each site (C2U mutations) and the proportion of C-to-U editing (U/C).
Supplementary Table 2: C-to-U editing events induced by APOBEC1 alone or APOBEC1-YTHmut.
Listed are the genome coordinates of DART-seq C-to-U editing sites identified from cells expressing APOBEC1-YTH. Shown are the C-to-U site coordinates and proportion of C-to-U editing at each site (U/C). Both high-stringency (C-to-U editing in at least 10% of total reads at each site) and low-stringency (C-to-U editing in at least 5% of total reads at each site) lists are included.
Supplementary Table 3: DART-seq sites in METTL3-depleted cells.
Listed are the genome coordinates of DART-seq C-to-U editing sites identified from HEK293T cells depleted of METTL3. Also indicated are the number of C-to-U mutations at each site (C2U mutations) and the proportion of C-to-U editing (U/C).
Supplementary Table 4: DART-seq sites identified from low-input samples
Listed are coordinates of DART-seq sites and the proportion of C-to-U editing at each site (U/C) for samples prepared from 10 ng or 100 ng of total RNA as input.
Supplementary Table 5: Differential gene expression analysis in APOBEC1-YTH-expressing cells.
Listed are genes which are up- or downregulated in HEK293T cells after 24 h of APOBEC1-YTH expression (n = 3 independent samples), APOBEC1-YTHmut expression (n = 3 independent samples) or APOBEC1 alone expression (n = 2 independent samples). Adjusted P < 0.05. Genes encoding major proteins in the m6A regulatory pathway are also highlighted.
Supplementary Table 6: mRNAs identified by in vitro and cellular DART-seq.
Listed are methylated mRNAs identified by in vitro DART-seq using cellular RNA and by cellular DART-seq using APOBEC1-YTH expression in HEK293T cells.
Supplementary Table 7: DART-seq sites altered by CPT treatment.
Genome coordinates of DART-seq sites identified after 5 h of camptothecin treatment. The proportion of C-to-U editing (U/C) at each site is also shown.
Supplementary Protocol
This supplementary protocol details the DART-seq method.
Source data
Rights and permissions
About this article
Cite this article
Meyer, K.D. DART-seq: an antibody-free method for global m6A detection. Nat Methods 16, 1275–1280 (2019). https://doi.org/10.1038/s41592-019-0570-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0570-0
This article is cited by
-
Decoding epitranscriptomic regulation of viral infection: mapping of RNA N6-methyladenosine by advanced sequencing technologies
Cellular & Molecular Biology Letters (2024)
-
Programmable protein expression using a genetically encoded m6A sensor
Nature Biotechnology (2024)
-
GEMS enables mRNA m6A methylation sensing in living cells and m6A-coupled protein delivery
Nature Biotechnology (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy
Signal Transduction and Targeted Therapy (2024)