Abstract
The bacterium Mycobacterium tuberculosis (Mtb) causes tuberculosis and is responsible for more human mortality than any other single pathogen1. Progression to active disease occurs in only a fraction of infected individuals and is predicted by an elevated type I interferon (IFN) response2,3,4,5,6,7. Whether or how IFNs mediate susceptibility to Mtb has been difficult to study due to a lack of suitable mouse models6,7,8,9,10,11. Here, we examined B6.Sst1S congenic mice that carry the ‘susceptible’ allele of the Sst1 locus that results in exacerbated Mtb disease12,13,14. We found that enhanced production of type I IFNs was responsible for the susceptibility of B6.Sst1S mice to Mtb. Type I IFNs affect the expression of hundreds of genes, several of which have previously been implicated in susceptibility to bacterial infections6,7,15,16,17,18. Nevertheless, we found that heterozygous deficiency in just a single IFN target gene, Il1rn, which encodes interleukin-1 receptor antagonist (IL-1Ra), is sufficient to reverse IFN-driven susceptibility to Mtb in B6.Sst1S mice. In addition, antibody-mediated neutralization of IL-1Ra provided therapeutic benefit to Mtb-infected B6.Sst1S mice. Our results illustrate the value of the B6.Sst1S mouse to model IFN-driven susceptibility to Mtb, and demonstrate that IL-1Ra is an important mediator of type I IFN-driven susceptibility to Mtb infections in vivo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data are available in the main text, Extended Data figures or the Supplementary Information.
Change history
16 April 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Global Tuberculosis Report 2018 (World Health Organization, 2018).
Zak, D. E. et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387, 2312–2322 (2016).
Scriba, T. J. et al. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 13, e1006687 (2017).
Berry, M. P. R. et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 (2010).
Singhania, A. et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat. Commun. 9, 2308 (2018).
Moreira-Teixeira, L., Mayer-Barber, K., Sher, A. & O’Garra, A. Type I interferons in tuberculosis: foe and occasionally friend. J. Exp. Med. 215, 1273–1285 (2018).
Donovan, M. L., Schultz, T. E., Duke, T. J. & Blumenthal, A. Type I interferons in the pathogenesis of tuberculosis: molecular drivers and immunological consequences. Front. Immunol. 8, 1633 (2017).
Stanley, S. A., Johndrow, J. E., Manzanillo, P. & Cox, J. S. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 178, 3143–3152 (2007).
Antonelli, L. R. V. et al. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J. Clin. Invest. 120, 1674–1682 (2010).
Desvignes, L., Wolf, A. J. & Ernst, J. D. Dynamic roles of type I and type II IFNs in early infection with Mycobacterium tuberculosis. J. Immunol. 188, 6205–6215 (2012).
Moreira-Teixeira, L. et al. Type I IFN inhibits alternative macrophage activation during Mycobacterium tuberculosis infection and leads to enhanced protection in the absence of IFN-γ signaling. J. Immunol. 197, 4714–4726 (2016).
Pan, H. et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature 434, 767–772 (2005).
Pichugin, A. V., Yan, B. S., Sloutsky, A., Kobzik, L. & Kramnik, I. Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am. J. Pathol. 174, 2190–2201 (2009).
Kramnik, I., Dietrich, W. F., Demant, P. & Bloom, B. R. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 97, 8560–8565 (2002).
Mayer-Barber, K. D. et al. Innate and adaptive interferons suppress IL-1α and IL-1β production by distinct pulmonary myeloid subsets during Mycobacterium tuberculosis infection. Immunity 35, 1023–1034 (2011).
Mayer-Barber, K. D. et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511, 99–103 (2014).
McNab, F. W. et al. Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-γ for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. J. Immunol. 193, 3600–3612 (2014).
Boxx, G. M. & Cheng, G. The roles of type I interferon in bacterial infection. Cell Host Microbe 19, 760–769 (2016).
Nunes-Alves, C. et al. In search of a new paradigm for protective immunity to TB. Nat. Rev. Microbiol. 12, 289–299 (2014).
Hawn, T. R., Shah, J. A. & Kalman, D. New tricks for old dogs: countering antibiotic resistance in tuberculosis with host-directed therapeutics. Immunol. Rev. 264, 344–362 (2015).
Zhang, G. et al. A proline deletion in IFNAR1 impairs IFN-signaling and underlies increased resistance to tuberculosis in humans. Nat. Commun. 9, 85 (2018).
De Oliveira Uehara, S. N. et al. High incidence of tuberculosis in patients treated for hepatitis C chronic infection. Braz. J. Infect. Dis. 20, 205–209 (2016).
Matsuoka, S. et al. Onset of tuberculosis from a pulmonary latent tuberculosis infection during antiviral triple therapy for chronic hepatitis C. Intern. Med. 55, 2011–2017 (2016).
Sabbatani, S. et al. Reactivation of severe, acute pulmonary tuberculosis during treatment with pegylated interferon-alpha and ribavirin for chronic HCV hepatitis. Scand. J. Infect. Dis. 38, 205–208 (2006).
Manca, C. et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl Acad. Sci. USA 98, 5752–5757 (2001).
Dorhoi, A. et al. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur. J. Immunol. 44, 2380–2393 (2014).
Teles, R. M. B. et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 339, 1448–1453 (2013).
McNab, F., Mayer-Barber, K., Sher, A., Wack, A. & O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015).
Bhattacharya, B. et al. The integrated stress response mediates type I interferon driven necrosis in Mycobacterium tuberculosis granulomas. Preprint at biorxiv.org/content/10.1101/499467v1 (2018).
He, X. et al. The sst1 resistance locus regulates evasion of type I interferon signaling by Chlamydia pneumoniae as a disease tolerance mechanism. PLoS Pathog. 9, e1003569 (2013).
Dunn, G. P. et al. A critical function for type I interferons in cancer immunoediting. Nat. Immunol. 6, 722–729 (2005).
Watson, R. O. et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17, 811–819 (2015).
Wassermann, R. et al. Mycobacterium tuberculosis differentially activates cGAS-and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe 17, 799–810 (2015).
Wiens, K. E. & Ernst, J. D. The mechanism for type I interferon induction by Mycobacterium tuberculosis is bacterial strain-dependent. PLoS Pathog. 12, 1–20 (2016).
Dey, B. et al. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 21, 401–408 (2015).
Collins, A. C. et al. Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host Microbe 17, 820–828 (2015).
Manzanillo, P. S., Shiloh, M. U., Portnoy, D. A. & Cox, J. S. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11, 469–480 (2012).
Eshleman, E. M., Delgado, C., Kearney, S. J., Friedman, R. S. & Lenz, L. L. Down regulation of macrophage IFNGR1 exacerbates systemic L. monocytogenes infection. PLoS Pathog. 13, 1–22 (2017).
Rayamajhi, M., Humann, J., Penheiter, K., Andreasen, K. & Lenz, L. L. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. J. Exp. Med. 207, 327–337 (2010).
Reboldi, A. et al. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science 345, 679–684 (2014).
Novikov, A. et al. Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1β production in human macrophages. J. Immunol. 187, 2540–2547 (2011).
Mayer-Barber, K. D. & Yan, B. Clash of the cytokine titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell. Mol. Immunol. 14, 22–35 (2017).
Mishra, B. B. et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat. Immunol. 14, 52–60 (2013).
Mishra, B. B. et al. Nitric oxide prevents a pathogen-permissive granulocytic inflammation during tuberculosis. Nat. Microbiol. 2, 17072 (2017).
Nichols, R. D., Von Moltke, J. & Vance, R. E. NAIP/NLRC4 inflammasome activation in MRP8+ cells is sufficient to cause systemic inflammatory disease. Nat. Commun. 8, 2209 (2017).
Fremond, C. M. et al. IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J. Immunol. 179, 1178–1189 (2007).
Mayer-Barber, K. D. et al. Caspase-1 independent IL-1beta production is critical for host resistance to Mycobacterium tuberculosis and does not require TLR signaling in vivo. J. Immunol. 184, 3326–3330 (2010).
Yamada, H., Mizumo, S., Horai, R., Iwakura, Y. & Sugawara, I. Protective role of interleukin-1 in mycobacterial infection in IL-1 alpha/beta double-knockout mice. Lab. Invest. 80, 759–767 (2000).
Sugawara, I., Yamada, H., Hua, S. & Mizuno, S. Role of interleukin (IL)-1 type 1 receptor in mycobacterial infection. Microbiol. Immunol. 45, 743–750 (2001).
Di Paolo, N. C. et al. Interdependence between interleukin-1 and tumor necrosis factor regulates TNF-dependent control of Mycobacterium tuberculosis infection. Immunity 43, 1125–1136 (2015).
Juffermans, N. P. et al. Interleukin‐1 signaling is essential for host defense during murine pulmonary tuberculosis. J. Infect. Dis. 182, 902–908 (2002).
Bohrer, A. C., Tocheny, C., Assmann, M., Ganusov, V. V. & Mayer-Barber, K. D. Cutting edge: IL-1R1 mediates host resistance to Mycobacterium tuberculosis by trans-protection of infected cells. J. Immunol. 201, 1645–1650 (2018).
Eisenberg, S. P. et al. Primary structure and functional expression from complementary DNA of a human interleukin-1 receptor antagonist. Nature 343, 341–346 (1990).
Dinarello, C. A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 281, 8–27 (2018).
Molnarfi, N., Hyka-Nouspikel, N., Gruaz, L., Dayer, J.-M. & Burger, D. The production of IL-1 receptor antagonist in IFN-beta-stimulated human monocytes depends on the activation of phosphatidylinositol 3-kinase but not of STAT1. J. Immunol. 174, 2974–2980 (2005).
Corr, M. et al. Interleukin 1 receptor antagonist mediates the beneficial effects of systemic interferon beta in mice: implications for rheumatoid arthritis. Ann. Rheum. Dis. 70, 858–863 (2011).
Cohen, S. B. et al. Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe 24, 439–446 (2018).
Hirsch, E., Irikura, V. M., Paul, S. M. & Hirsh, D. Functions of interleukin 1 receptor antagonist in gene knockout and overproducing mice. Proc. Natl Acad. Sci. USA 93, 11008–11013 (2002).
Nicklin, M. J. H., Hughes, D. E., Barton, J. L., Ure, J. M. & Duff, G. W. Arterial inflammation in mice lacking the interleukin 1 receptor antagonist gene. J. Exp. Med. 191, 303–312 (2000).
Fujioka, N. et al. Preparation of specific antibodies against murine IL-1ra and the establishment of IL-1ra as an endogenous regulator of bacteria-induced fulminant hepatitis in mice. J. Leukoc. Biol. 58, 90–98 (1995).
Lei, X., Zhu, H., Zha, L. & Wang, Y. SP110 gene polymorphisms and tuberculosis susceptibility: a systematic review and meta-analysis based on 10 624 subjects. Infect. Genet. Evol. 12, 1473–1480 (2012).
Cai, L. et al. Identification of genetic associations of SP110/MYBBP1A/RELA with pulmonary tuberculosis in the Chinese Han population. Hum. Genet. 132, 265–273 (2013).
Fox, G. J. et al. Polymorphisms of SP110 are associated with both pulmonary and extra-pulmonary tuberculosis among the Vietnamese. PLoS ONE 9, e99496 (2014).
Ren, G. et al. SP110 and PMP22 polymorphisms are associated with tuberculosis risk in a Chinese-Tibetan population. Oncotarget 7, 66100–66108 (2016).
Leu, J. S. et al. SP110b controls host immunity and susceptibility to tuberculosis. Am. J. Respir. Crit. Care Med. 195, 369–382 (2017).
Zhang, S. et al. Certain polymorphisms in SP110 gene confer susceptibility to tuberculosis: a comprehensive review and updated meta-analysis. Yonsei Med. J. 58, 165–173 (2017).
Zhou, Y. et al. Polymorphisms in the SP110 and TNF-α gene and susceptibility to pulmonary and spinal tuberculosis among southern Chinese population. Dis. Markers 2017, 4590235 (2017).
Chang, S.-Y. et al. SP110 polymorphisms are genetic markers for vulnerability to latent and active tuberculosis infection in Taiwan. Dis. Markers 2018, 4687380 (2018).
Mitnick, C. D. et al. Aggressive regimens for multidrug-resistant tuberculosis decrease all-cause mortality. PLoS ONE 8, e58664 (2013).
Chung-Delgado, K., Guillen-Bravo, S., Revilla-Montag, A. & Bernabe-Ortiz, A. Mortality among MDR-TB cases: comparison with drug-susceptible tuberculosis and associated factors. PLoS ONE 10, e0119332 (2015).
Bauman, D. R. et al. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc. Natl Acad. Sci. USA 106, 16764–16769 (2009).
Sauer, J.-D. et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011).
Acknowledgements
We thank the Stanley and Cox laboratories for discussions and for support with Mtb experiments, L. Flores, P. Dietzen and R. Chavez for technical assistance, and H. Nolla, A. Valeros and the Cancer Research Laboratory for flow cytometry. We thank the Barton laboratory, I. Rauch, A. Sandstrom, D. Kotov and P. Mitchell for helpful discussions and technical support. We thank B. Penn for comments on the manuscript. Generation of the anti-IL-1Ra antibody was supported by the Extramural Collaborative Research Program of the Cancer Research Institute, Kanazawa University. K.H.D. and R.E.V. were supported by Investigator in the Pathogenesis of Infectious Diseases awards from the Burroughs Wellcome Fund. R.E.V. is an HHMI Investigator and is supported by NIH grants AI075039 and AI066302.
Author information
Authors and Affiliations
Contributions
R.E.V., K.H.D. and D.X.J. designed the experiments. L.H.Y. assisted with experiments shown in Fig. 4 and Extended Data Fig. 5. K.J.C. assisted with experiments shown in Extended Data Fig. 3. D.X.J. performed all other experiments. R.E.V. and D.X.J. analysed the data. I.K. generated the B6.Sst1S mice. N.M. generated the anti-IL-1Ra antibody. K.H.D. and I.K. gave technical support and conceptual advice. R.E.V. and D.X.J. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.E.V has a financial relationship with Aduro Biotech and both he and the company may benefit from commercialization of the results of this research. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
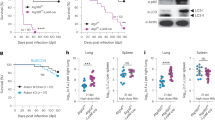
Extended Data Fig. 1 B6.Sst1S BMMs overexpress Ifnb and ISGs when stimulated with TNFα.
a, b, Expression of Ifnb (a) or selected ISGs (b) in BMMs measured by RT-qPCR. Results normalized to housekeeping genes. Representative data of at least two independent experiments.
Extended Data Fig. 2 B6.Sst1SStinggt/gt partially rescues the enhance susceptibility of B6.Sst1S mice to Mtb.
Mice were infected with Mtb and measured for lung bacterial burdens at day 25 (a) or survival (b). a, combined results of 2 experiments. Sample size n (B6, B6.Sst1S, B6.Sst1SStinggt/gt) = 11, 11, 12 (a); 11, 11, 11 (b). All animals except 5 of the B6 were bred in-house (a) and all except B6 were bred in-house (b). Center and error bars show mean and SEM. Analyzed with two-ended Mann–Whitney test (a) or two-ended Log-rank test (b). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
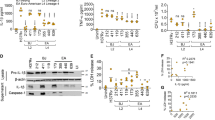
Extended Data Fig. 3 Enhanced inflammation in B6.Sst1S mice requires type I IFN.
a, b, e, f, Protein levels of IL-10 (a), IFNγ (b), TNF (e), and CXCL1 (f) were measured in lungs of Mtb-infected mice at day 25. Combined results of at least three independent infections (a, b, e, f). c, Lung bacterial burden of Mtb-infected mice at day 25 (representative of two independent infections). Input dose: average 100 CFU/mouse. d, CFU corresponding to Fig. 2a and b. Combined results that include those already shown in Figs. 1c, 2c, and Extended Data 3a. Input dose: 10–89 CFU per mouse. g, Neutrophils (CD11b+Ly6G+) from lungs of Mtb-infected mice were enumerated on day 14 and day 25. Combined results of two independent infections. All animals except B6 were bred in-house (a–f); all animals were bred in-house (g). Sample size n (B6, B6.Sst1S, B6.Sst1SIfnar–/–) = 16, 18, 16 (a); 40, 45, 32 (b); 40, 44, 32 (d); 25, 29, 16 (e); 28, 30, 26 (f); 6 for all genotypes (c); 9 for all genotypes (g). Center and error bars show mean and SEM. Analyzed with two-ended Mann–Whitney test (a–g). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Extended Data Fig. 4 IL-1 blockade increases susceptibility in both B6 and B6.Sst1S mice.
a–c, mice were infected with average 15 CFU/mouse and were treated with anti-IL1R1 or isotype control antibodies, and on day 25 the lungs were measured for bacterial burden (a), IL- 1Ra protein levels (b), and IL-1 bioactivity (c). Sample size n (in order shown, from left) = 7, 7, 8, 8 (a); 7, 8, 9, 8 (b-c). d, Mice were infected with average 78 CFU/mouse and IL-1 bioactivity was measured in the lungs at day 25. n = 5 for both samples. e, Mice were infected with average 33 CFU/mouse. IL-1 bioactivity was measured in lung samples collected on the indicated days. n = 6 for all samples except day 36 B6.Sst1S = 4. Day 25 data already shown in Fig. 3d. All animals except B6 were bred in-house (a–c); all were bred in-house (d-e). Center and error bars show mean and SEM. Analyzed with two-ended Mann–Whitney test (a–c). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Extended Data Fig. 5 Homozygous or heterozygous Il1rn deletion protects B6.Sst1S mice from Mtb infection.
a, Body weights on day 28 of individual mice shown in Fig. 4c. b-c, Mice were infected with Mtb at average 17 CFU/mouse (b) or 60 CFU/mouse (c), and at day 21 lungs were harvested to measure IL-1 bioactivity. d, RT-qPCR on lungs of Mtb-infected mice, sampled at 25 days post-infection. Each graph combined results from 2 independent experiments. e-f, Mice infected with average 20 CFU/mouse were treated with either anti-IL-1R1 antibody or isotype control every 3 days starting 7 days post-infection (e), or anti-IL-1Ra antibody or PBS control every other day starting 3 days post-infection (f). At 25 days post-infection lungs were harvested for measuring bacterial burden. Sample size n (from left as shown) = 11, 12, 22, 10 (a); 6, 8, 11 (b); 6, 8, 10 (c); 11, 14, 11, 5(d); 5, 6, 3, 4, 2, 3 (e); 8, 8, 6, 6 (f). All mice were bred in-house (a, d–f) or all except B6 were bred in-house (b-c); and all except B6 and B6.Sst1S were littermates (a, d, e). Center and error bars show mean and SEM. Analyzed with two-ended Mann–Whitney test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Ji, D.X., Yamashiro, L.H., Chen, K.J. et al. Type I interferon-driven susceptibility to Mycobacterium tuberculosis is mediated by IL-1Ra. Nat Microbiol 4, 2128–2135 (2019). https://doi.org/10.1038/s41564-019-0578-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0578-3
This article is cited by
-
Multiple cytokine analysis based on QuantiFERON-TB gold plus in different tuberculosis infection status: an exploratory study
BMC Infectious Diseases (2024)
-
Fecal microbiota impacts development of Cryptosporidium parvum in the mouse
Scientific Reports (2024)
-
From immunology to artificial intelligence: revolutionizing latent tuberculosis infection diagnosis with machine learning
Military Medical Research (2023)
-
Host-directed immunotherapy of viral and bacterial infections: past, present and future
Nature Reviews Immunology (2023)
-
ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages
Nature Microbiology (2023)