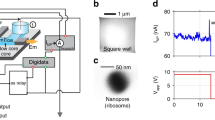
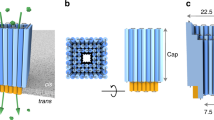
Abstract
The precise assembly and engineering of molecular machines capable of handling biomolecules play crucial roles in most single-molecule methods. In this work we use components from all three domains of life to fabricate an integrated multiprotein complex that controls the unfolding and threading of individual proteins across a nanopore. This 900 kDa multicomponent device was made in two steps. First, we designed a stable and low-noise β-barrel nanopore sensor by linking the transmembrane region of bacterial protective antigen to a mammalian proteasome activator. An archaeal 20S proteasome was then built into the artificial nanopore to control the unfolding and linearized transport of proteins across the nanopore. This multicomponent molecular machine opens the door to two approaches in single-molecule protein analysis, in which selected substrate proteins are unfolded, fed to into the proteasomal chamber and then addressed either as fragmented peptides or intact polypeptides.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All relevant data are included in the article and its Supplementary Information. Statistical source data, unmodified gels, and molecular dynamics simulations results are provided in Source data. Data is also available from the authors upon reasonable request. Source data are provided with this paper.
References
Bayley, H. Nanopore sequencing: from imagination to reality. Clin. Chem. 61, 25–31 (2014).
Kang, X. F., Cheley, S., Guan, X. & Bayley, H. Stochastic detection of enantiomers. J. Am. Chem. Soc. 128, 10684–10685 (2006).
Huang, G., Voet, A. & Maglia, G. FraC nanopores with adjustable diameter identify the mass of opposite-charge peptides with 44 dalton resolution. Nat. Commun. 10, 1–10 (2019).
Huang, G., Willems, K., Soskine, M., Wloka, C. & Maglia, G. Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun. 8, 935 (2017).
Restrepo-Pérez, L., Wong, C. H., Maglia, G., Dekker, C. & Joo, C. Label-free detection of post-translational modifications with a nanopore. Nano Lett. 19, 7957–7964 (2019).
Ouldali, H. et al. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 38, 176–181 (2020).
Hu, Z.-L., Huo, M.-Z., Ying, Y.-L. & Long, Y.-T. Biological nanopore approach for single‐molecule protein sequencing. Angew. Chemie 60, 14738–14749 (2020).
Nivala, J., Mulroney, L., Li, G., Schreiber, J. & Akeson, M. Discrimination among protein variants using an unfoldase-coupled nanopore. ACS Nano 8, 12365–12375 (2014).
Nivala, J., Marks, D. B. & Akeson, M. Unfoldase-mediated protein translocation through an α-hemolysin nanopore. Nat. Biotechnol. 31, 247–250 (2013).
Xu, C. et al. Computational design of transmembrane pores. Nature 585, 129–134 (2020).
Joh, N. H. et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1520 (2014).
Lu, P. et al. Accurate computational design of multipass transmembrane proteins. Science 359, 1042–1046 (2018).
Scott, A. et al. Constructing ion channels from water-soluble α-helical barrels. Nat. Chem. 13, 643–650 (2021).
Spruijt, E., Tusk, S. E. & Bayley, H. DNA scaffolds support stable and uniform peptide nanopores. Nat. Nanotechnol. 13, 739–745 (2018).
Seemüller, E. et al. Proteasome from Thermoplasma acidophilum: a threonine protease. Science 268, 579–582 (1995).
Löwe, J. et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science 268, 533–539 (1995).
Sugiyama, M. et al. Spatial arrangement and functional role of α subunits of proteasome activator PA28 in hetero-oligomeric form. Biochem. Biophys. Res. Commun. 432, 141–145 (2013).
Förster, A., Masters, E. I., Whitby, F. G., Robinson, H. & Hill, C. P. The 1.9 Å structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 18, 589–599 (2005).
Jiang, J., Pentelute, B. L., Collier, R. J. & Hong Zhou, Z. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature 521, 545–549 (2015).
Cheley, S., Braha, O., Lu, X., Conlan, S. & Bayley, H. A functional protein pore with a "retro" transmembrane domain. Protein Sci. 8, 1257–1267 (1999).
Gu, L. Q. et al. Reversal of charge selectivity in transmembrane protein pores by using noncovalent molecular adapters. Proc. Natl Acad. Sci. USA 97, 3959–3964 (2000).
Maglia, G., Restrepo, M. R., Mikhailova, E. & Bayley, H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc. Natl Acad. Sci. USA 105, 19720–19725 (2008).
Chen, B. et al. Engagement of arginine finger to ATP triggers large conformational changes in NtrC1 AAA+ ATPase for remodeling bacterial RNA polymerase. Structure 18, 1420–1430 (2010).
Gu, L. Q., Braha, O., Conlan, S., Cheley, S. & Bayley, H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature 398, 686–690 (1999).
Yannakopoulou, K. et al. Symmetry requirements for effective blocking of pore-forming toxins: comparative study with α-, β-, and γ-cyclodextrin derivatives. Antimicrob. Agents Chemother. 55, 3594–3597 (2011).
Förster, A. & Hill, C. P. Proteasome activators. Protein Degrad. 2, 89–110 (2007).
Huber, E. M. & Groll, M. The mammalian proteasome activator PA28 forms an asymmetric α4β3 complex. Structure 25, 1473–1480.e3 (2017).
Kuehn, L. & Dahlmann, B. Proteasome activator PA28 and its interaction with 20S proteasomes. Arch. Biochem. Biophys. 329, 87–96 (1996).
Benaroudj, N., Zwickl, P., Seemüller, E., Baumeister, W. & Goldberg, A. L. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 11, 69–78 (2003).
Huang, R. et al. Unfolding the mechanism of the AAA+ unfoldase VAT by a combined cryo-EM, solution NMR study. Proc. Natl Acad. Sci. USA 113, E4090–W4199 (2016).
Ripstein, Z. A., Huang, R., Augustyniak, R., Kay, L. E. & Rubinstein, J. L. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife 6, 1–14 (2017).
Gerega, A. et al. VAT, the Thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J. Biol. Chem. 280, 42856–42862 (2005).
Akopian, T. N., Kisselev, A. F. & Goldberg, A. L. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J. Biol. Chem. 272, 1791–1798 (1997).
Pédelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Ward, W. W., Prentice, H. J., Roth, A. F., Cody, C. W. & Reeves, S. C. Spectral perturbations of the aequorea green-fluorescent protein. Photochem. Photobiol. 35, 803–808 (1982).
Hsu, S.-T. D., Blaser, G. & Jackson, S. E. The folding, stability and conformational dynamics of β-barrel fluorescent proteins. Chem. Soc. Rev. 38, 2951–2965 (2009).
Kisselev, A. F., Songyang, Z. & Goldberg, A. L. Why does threonine, and not serine, function as the active site nucleophile in proteasomes? J. Biol. Chem. 275, 14831–14837 (2000).
Biesemans, A., Soskine, M. & Maglia, G. A protein rotaxane controls the translocation of proteins across a ClyA nanopore. Nano Lett. 15, 6076–6081 (2015).
Majumder, P. et al. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc. Natl Acad. Sci. USA 116, 534–539 (2019).
Kisselev, A. F., Akopian, T. N. & Goldberg, A. L. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J. Biol. Chem. 273, 1982–1989 (1998).
Maglia, G., Heron, A. J. J., Stoddart, D., Japrung, D. & Bayley, H. Analysis of single nucleic acid molecules with protein nanopores. Methods Enzym. 475, 591–623 (2010).
Acknowledgements
This work is financially supported by ERC consolidator grant (no. 726151).
Author information
Authors and Affiliations
Contributions
S.Z. and G.M. designed the experiments. G.M. supervised the project. S.Z. performed the experiments and data analysis. B.M.H.B., P.C.T.d.S. and S.-J.M. conducted the simulation work. G.M. and S.Z. wrote the paper. All authors discussed the results, and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
G.M. is a founder, director and shareholder of Portal Biotech Limited, a company engaged in the development of nanopore technologies. This work was not supported by Portal Biotech Limited.
Additional information
Peer review information Nature Chemistry thanks Ulrich Keyser, Yi-Lun Ying and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Additional discussion, methods, Supplementary Figs. 1–29, Table 1 and references.
Supplementary Data 1
molecular dynamics simulations of REG–nanopore for Fig. 1d.
Supplementary Data 2
molecular dynamics simulations of proteasome–nanopore for Fig. 1h.
Supplementary Data 3
Statistical Source Data for Supplementary Fig. 7.
Supplementary Data 4
Statistical Source Data for Supplementary Fig. 28.
Supplementary Data
Unprocessed gels.
Source data
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Unprocessed gels.
Source Data Fig. 5
Statistical Source Data.
Rights and permissions
About this article
Cite this article
Zhang, S., Huang, G., Versloot, R.C.A. et al. Bottom-up fabrication of a proteasome–nanopore that unravels and processes single proteins. Nat. Chem. 13, 1192–1199 (2021). https://doi.org/10.1038/s41557-021-00824-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00824-w
This article is cited by
-
Real-time detection of 20 amino acids and discrimination of pathologically relevant peptides with functionalized nanopore
Nature Methods (2024)
-
Machine learning for functional protein design
Nature Biotechnology (2024)
-
Nanopore DNA sequencing technologies and their applications towards single-molecule proteomics
Nature Chemistry (2024)
-
Peptide sequencing based on host–guest interaction-assisted nanopore sensing
Nature Methods (2024)
-
Unambiguous discrimination of all 20 proteinogenic amino acids and their modifications by nanopore
Nature Methods (2024)