Abstract
A taxonomic classification that accurately captures evolutionary history is essential for conservation. Genomics provides powerful tools for delimiting species and understanding their evolutionary relationships. This allows for a more accurate and detailed view on conservation status compared with other, traditionally used, methods. However, from a practical and ethical perspective, gathering sufficient samples for endangered taxa may be difficult. Here, we use museum specimens to trace the evolutionary history and species boundaries in an Asian oriole clade. The endangered silver oriole has long been recognized as a distinct species based on its unique coloration, but a recent study suggested that it might be nested within the maroon oriole-species complex. To evaluate species designation, population connectivity, and the corresponding conservation implications, we assembled a de novo genome and used whole-genome resequencing of historical specimens. Our results show that the silver orioles form a monophyletic lineage within the maroon oriole complex and that maroon and silver forms continued to interbreed after initial divergence, but do not show signs of recent gene flow. Using a genome scan, we identified genes that may form the basis for color divergence and act as reproductive barriers. Taken together, our results confirm the species status of the silver oriole and highlight that taxonomic revision of the maroon forms is urgently needed. Our study demonstrates how genomics and Natural History Collections (NHC) can be utilized to shed light on the taxonomy and evolutionary history of natural populations and how such insights can directly benefit conservation practitioners when assessing wild populations.
Similar content being viewed by others
Introduction
With species and populations disappearing at an unprecedented rate (Raven and Miller 2020), accurately characterizing global biodiversity is becoming increasingly urgent. Species designation and evolutionary history are crucial aspects to be considered in conservation because this knowledge helps to assess the degree of isolation between species, identify unique lineages that preserve critical genetic diversity, and optimize management plans to maximize adaptive potential. As such, the fields of taxonomy and systematics, which have been largely based on phenotypic traits and/or small sets of genetic markers, are fundamental for the protection of biodiversity. However, with a growing ability to characterize genome-wide variation, we have become increasingly aware that single gene trees do not always align with the species tree (Maddison 1997) and that genetic divergence does not always scale with phenotypic differentiation (Dussex et al. 2018). While some distant species pairs can be indiscernible in terms of morphology, populations that do exhibit phenotypic differences can lack notable genetic differentiation (Rheindt et al. 2011). Slight changes to small portions of the genome can have a large phenotypic effect, as demonstrated by the increasing number of genome-wide studies that show high levels of genetic homogeneity between species displaying striking differences in plumage coloration (Poelstra et al. 2014; Toews et al. 2016; Campagna et al. 2017). Discordance between genetic and phenotypic data can have severe implications for conservation, since a taxonomic classification that under- or overestimates genetic divergence leads to inappropriate delineation and prioritization of conservation units (Stanton et al. 2019).
High-throughput sequencing is now a well-established tool in conservation science (McMahon et al. 2014) and genomics offers powerful ways to characterize the patterns and evolutionary processes shaping lineage diversity (Supple and Shapiro 2018). With sequencing costs having decreased substantially over the past decade (Schloss et al. 2020), empiricists are no longer limited by sequencing capacity. Instead, access to a representative collection of samples, that covers geographic spread and intraspecific phenotypic variation, can be a major limiting factor and is particularly challenging for species of conservation concern (Blom 2021). Endangered species are frequently elusive and have small population sizes. This often impedes the sampling needed to study demography, biogeography, and population connectivity (Thompson 2013). Furthermore, endangered species tend to be vulnerable to human impact and, as such, the use of invasive sampling poses ethical challenges (Walters 2006; Winker et al. 2010; Costello et al. 2016). Therefore, while the technology may be in place, studying species of conservation concern remains a challenging and costly exercise that is frequently accompanied by ethical considerations.
Natural History Collections (NHCs) are important repositories of biodiversity across time and space, and represent a unique resource for the study of endangered taxa (Payne and Sorenson 2002; Wandeler et al. 2007; Bi et al. 2013; Holmes et al. 2016). Furthermore, historic specimens are testimonies of past life on earth and therefore important references to which contemporary specimens can be compared in order to understand how recent disturbance has affected wild populations. For instance, they have been successfully used to detect temporal trends in demography and genetic diversity (Miller and Waits 2003; Mondol et al. 2013). However, a major drawback of historical samples is that their DNA often suffers from postmortem DNA fragmentation and damage (Burrell et al. 2015), which can complicate the application of genomic methods. To analyze whole-genome datasets, researchers strongly rely on existing genomic resources such as high-quality genome assemblies to which degraded DNA can be compared. Even though such resources have remained largely absent for many taxa, recent developments make it increasingly tractable to generate reference genomes de novo, which permits the wide-scale utilization of NHCs in the study of endangered taxa. Here, we use linked-read sequencing to assemble a de novo reference genome and use whole-genome resequencing of historical specimens to study the evolutionary history and taxonomic status of an avian species complex from Asia.
The endangered silver oriole, Oriolus mellianus, is a migratory songbird of the family Oriolidae and known to breed only in a few localities in South-Central China. Its wintering grounds are poorly known, with some records from Thailand and a few records from South-West Cambodia. Loss and fragmentation of forest habitat throughout the species’ range is likely responsible for the declining trajectory of this species, which today has an estimated population size between 1000 and 2499 individuals (BirdLife International 2016). Based on morphology, O. mellianus has been considered sister to the polytypic maroon oriole, O. traillii, with an extensive distribution across South-East Asia (Fig. 1a). Male individuals display striking phenotypic differences, as O. mellianus specimens are coated silvery white in body parts where O. traillii has different hues of crimson maroon (Fig. 1b). Despite these plumage differences, recent phylogenetic analyses based on a few genetic markers have suggested that O. mellianus may be nested within O. traillii and perhaps even within the geographically adjacent subspecies O. t. traillii (Jønsson et al. 2019). However, the study by Jønsson et al. (2019) included a single O. mellianus individual, making it challenging to discriminate between competing hypotheses that could explain the observed branching patterns, such as recent phenotypic divergence or hybridization between distinct morphological forms.
a Map showing the breeding ranges of O. mellianus and the O. traillii subspecies (O. t. traillii, O. t. robinsoni, O. t. nigellicauda, and O. t. ardens) according to BirdLife International (2016, 2018). Colored dots represent sampling locations and are tagged with their corresponding sample ID. The arrows depict estimated migration routes of O. mellianus. b Illustrations representing variations in plumages of O. traillii (upper) and O. mellianus (lower). c Best-scoring maximum-likelihood tree obtained from the concatenated nuclear sequences. The scale bar is measured in absolute divergence-time units. Taxa are colored according to the colors assigned in the distribution map.
Population-wide sampling and whole-genome sequencing provides a more detailed view on the evolutionary history and taxonomy of this species group. As these genomic resources were previously nonexisting, we generated the first reference assembly and whole-genome population-level dataset for members of the Oriolidae family. This allowed us to assess if O. mellianus represents a phylogenetically valid species and establish the systematic affinities of O. mellianus relative to other populations of O. traillii. Finally, we quantified the extent of ancient and extant population connectivity and searched for genomic regions of differentiation to locate candidate genes of importance for the development of the distinct plumage characters of O. mellianus. These results have direct implications for the conservation management and status of Asian orioles, underline how modern genomic approaches can provide a genome-wide view on population connectivity, and how NHCs represent a powerful resource for the study of endangered taxa.
Materials and methods
Sample collection, DNA extraction, library preparation and sequencing
No high-quality genome assembly was available for any oriole species. Thus, we assembled a de novo genome to use as a reference for the whole-genome resequencing data sourced from museum specimens. For the de novo genome assembly, we extracted DNA from a blood sample taken from a closely related black-hooded oriole (Oriolus xanthornus subsp. maderasperatus) individual sampled in India (ZMUC139617). DNA was extracted using the KingFisher DNA extraction platform. Subsequently, a linked-read sequencing library was prepared using the Chromium Genome platform (10X Genomics) and sequenced on the HiSeqX at SciLifeLab Uppsala (Sweden).
Furthermore, we generated whole-genome resequencing data for sixteen museum specimens collected between 1906 and 1952: six O. mellianus and ten O. traillii individuals, representing all currently recognized maroon oriole subspecies (five O. t. traillii, three O. t. robinsoni samples, one O. t. nigellicauda, and one O. t. ardens; Table S1). Based on geographic location and collection availability, we selected samples collected across South-East Asia (Fig. 1a). We sourced genetic material from toepads of museum-study skins, extracted DNA following the protocol described in Irestedt et al. (2006), and prepared four Illumina libraries per individual following the protocol published by Meyer and Kircher (2010). For each sample, we pooled the libraries in equimolar ratios before sending them to SciLifeLab Stockholm (Sweden) for paired-end sequencing (read length: 150 bp) using Illumina Next Generation Sequencing technology on the HiSeqX or NovaSeq sequencing platform.
De novo 10X chromium assembly
For the assembly of the O. xanthornus draft genome, the NextFlow-core pipeline Neutronstar was used. This is a best-practice assembly pipeline for the de novo assembly and quality control of 10x Genomics Chromium data. 10X Genomics linked-reads were assembled using Supernova, and in order to assess assembly quality and completeness, standard quality metrics were computed using QUAST (V5.0.2; Gurevich et al. 2013). In addition, the presence, completeness, and copy number of Benchmarking Universal Single-Copy Orthologs (BUSCOs) was evaluated using BUSCO (V3.0.2; Simão et al. 2015). For evaluation, the output of these assessment tools was visualized in a MultiQC (v1.7; Ewels et al. 2016) report.
Cleaning
Following sequencing, we polished the raw resequencing data using a custom-designed workflow for museum specimens (https://github.com/MozesBlom/NGSdata_tools/). This workflow consists of (i) removal of duplicates using Super-Deduper (V1.0.0; Petersen et al. 2015), (ii) adapter trimming using Trimmomatic (V0.36; Bolger et al. 2014), (iii) merging of overlapping read pairs with PEAR (V0.9.10; Zhang et al. 2014a), (iv) quality trimming with Trimmomatic (V0.36), and (v) removal of low-complexity reads using an in-house-developed Python script. Before and after the cleaning, we visually inspected the quality of the reads using FastQC (V0.11.5; Andrews 2010).
Mitogenome reconstruction
By blasting the whole mitogenome of O. chinensis (accession number: JQ083495) against the de novo genome assembly of O. xanthornus with Blastn (Zhang et al. 2000), we identified a single contig (ID 118416) scoring the complete mitogenome. After visually inspecting and curating this full O. xanthornus mitogenome sequence using Geneious (V2019.1.3; Kearse et al. 2012), we used it as a reference for reconstructing the mitogenomes of all focal individuals. Mitogenomes were reconstructed from the cleaned read data using MITObim (V1.9; Hahn et al. 2013), which uses an iterative baiting and mapping strategy and has been shown to effectively reconstruct mitogenomes even when there is no reference sequence for the target species (Hahn et al. 2013). After reconstructing the mitogenomes, the control region was excluded because of its complex evolutionary properties that hamper its reconstruction (Howell et al. 2007).
Mitochondrial phylogeny
Because of the linked nature of the mitogenome, we treated it as a single hereditary unit for phylogenetic inference. We selected the best-scoring tree out of 10 maximum-likelihood tree searches in RAxML (V8.2.10; Stamatakis 2014) and calculated the bootstrap support for each node with 1000 bootstrap replicates. For the phylogenetic inference, we selected the general time reversible (GTR) model of evolution with a gamma-distributed rate variation among sites based on the Bayesian information criterion provided by MEGA X (Kumar et al. 2018).
Mapping
In order to map the cleaned reads to the O. xanthornus reference genome, we used the BWA-MEM algorithm (V0.7.17; Li 2013). Then, we used SAMtools (V0.1.19; Li et al. 2009) to (i) convert the SAM files into BAM format, (ii) remove reads that did not pair properly with the reference (iii), sort and index the BAM files, and (iv) merge the library-specific BAM files by individual. Since short scaffolds have a higher likelihood to include assembly errors and are frequently repetitive in nature, we filtered each BAM file to exclusively keep reads mapping to the 1000 longest scaffolds (representing ~85% of the total genome size) for downstream analyses. The shortest scaffold included for downstream analyses spanned 41,402 bp. Last, we evaluated DNA damage in the final BAM files by using mapDamage (V2.0; Jónsson et al. 2013).
Variant calling and filtering
We performed variant calling and filtering using two alternative workflows based either on “hard-calling” genotypes or on inferring genotype likelihoods (GLs). The latter are able to incorporate statistical uncertainty through downstream analyses and recommended when working with low-coverage data, as is typically the case with historical samples (Korneliussen et al. 2014). For the GL approach, we used ANGSD (V0.921; Korneliussen et al. 2014) to jointly call biallelic SNPs. GLs were estimated using the GATK model (McKenna et al. 2010). For that calculation, we considered only bases with a quality score of at least 20 from reads that have a minimum mapping score of 20 and mapped to unique positions. Furthermore, we set the SNP p-value threshold to 1e–6, which has been reported as a conservative value (Korneliussen et al. 2014) and rejected sites with a significant p-value for being triallelic. Similarly, we discarded sites that had an individual coverage below 10 or above 100 reads, missing data for two or more individuals or a minor allele frequency (MAF) below 0.12. While the latter may lead to the removal of subspecies-specific alleles in O. t. ardens and O. t. nigellicauda, we decided for a stringent MAF threshold because we are dealing with medium-coverage data that show degradation patterns (Table S1; see below). In order to perform downstream analyses (NGSadmix and PCAngsd) on different sample subsets, we ran ANGSD twice: once including the total sample pool and once including only O. mellianus, O. t. robinsoni, and O. t. traillii, which we henceforth refer to as continental taxa.
For the hard-calling pipeline, we employed FreeBayes (V1.0.2; Garrison and Marth 2012) using the joint genotyping model with a population prior. Similar to the previous approach, genotypes were called exclusively considering bases with a phred quality score above 20 from reads with a mapping quality score above 20. Subsequently, we applied analogous filtering steps to the previous approach using VCFtools (V0.1.15; Danecek et al. 2011) and Vcflib (Garrison 2012). More specifically, we filtered variants by (i) eliminating individual genotypes if the depth was less than 10 or more than 100 reads per individual, (ii) removing sites if more than a single individual had missing data, (iii) decomposing complex variants into SNPs and indels before removing all variants that were not SNPs, (iii) eliminating nonbiallelic sites (including the reference allele), (iv) removing variants with more than one missing individual, and (v) dismissing variants with a MAF below 0.12 so that at least 4 chromosomes had to support the minor allele.
Phylogenetic framework and divergence-time estimation
We employed two distinct approaches to infer a nuclear phylogeny: a maximum-likelihood approach based on a genome-wide concatenated dataset and a full-coalescent approach based on a subset of unlinked biallelic SNPs. In order to infer the nuclear concatenated tree, we first generated consensus sequences for each individual based on the filtered VCF files by using BCFtools (V1.3.1; Li and Durbin 2009). Heterozygous sites were included and coded according to the IUPAC-ambiguity code. However, we masked sites if the coverage was below 10 reads or above three times the mean coverage of the individual under consideration. This was done by computing read depth at each position using SAMtools (V0.1.19) and subsequently masking the individual consensus sequences using BEDtools (V2.27.1; Quinlan and Hall 2010). Next, we extracted 10 kb windows in a 90 kb distance if the contig length exceeded 100 kb. For each window to be included, at least 50% of the positions had to have sequence data for at least 50% of all individuals. Ultimately, we based the phylogenetic inference on a concatenation of 9738 (10 kb) windows that passed these requirements. Then, we inferred a phylogeny by performing 20 maximum-likelihood tree searches using 10 random and 10 parsimony-based starting trees. These inferences were done under the GTR model of evolution for unphased diploid genotype data and default settings. In addition, we performed nonparametric bootstrap by resampling alignment columns and reinferring the tree for 100 replicates. We then computed branch-support values onto the best-scoring tree. Furthermore, we used Twisst (Martin and Van Belleghem 2017) to quantify the frequency of competing subtree topologies and estimate the number of windows supporting different phylogenies. The input file contained gene trees constructed based on the windows selected for the concatenated tree inference. For each window alignment, we performed a maximum-likelihood tree search using RAxML-NG (Kozlov et al. 2019) under the GTR model of evolution for unphased diploid genotype data. Last, in order to estimate divergence times, we calculated uncorrected sequence divergences (p-distances) between the samples from the concatenated dataset using DiStats (Astrin et al. 2016). Subsequently, we scaled the sequence-divergence estimates with an empirically informed mutation rate for birds (Zhang et al. 2014b; Smeds et al. 2016) to approximate the pairwise divergence times between all individuals (number of generations, Table S2).
Since concatenated analyses do not account for incomplete lineage sorting (ILS), we also used SNAPP (Bryant et al. 2012) to infer the species-tree under a full-coalescent model. This approach assumes that the loci are biallelic and unlinked. Whereas the former requirement is already met due to our filtering criteria, we thinned the variants by setting the minimum separation criterion between SNP positions to 10 kb in order to minimize linkage disequilibrium (Christmas et al. 2021). Next, we converted de VCF file into a binary nexus matrix using a custom python script (Ortiz 2019). The final SNAPP input file contained 84,236 SNPs. We used Beauti to generate a BEAST XML file in which we (i) assigned each individual to its respective population based on the results obtained by NGSadmix and PCAngsd (see below), (ii) fixed coalescence rate at 10 and forward and reverse mutation-rate parameters to 1, (iii) set the chain length for the MCMC to 1*10-e7 generations, and (iv) sampled trees every 1000th iteration. After the run, we discarded the first 10% of the chain as burn-in using TRACER (V1.7.1; Rambaut et al. 2018) and we assumed convergence of parameter estimates if the respective effective sample size was greater than 200.
Testing for hybridization
We took three alternative approaches to evaluate whether the observed incongruence between phenotypic and genetic similarity was caused by (a) recent speciation and phenotypic divergence or (b) hybridization between distinct morphological forms. First, we used PCAngsd (V0.98; Meisner and Albrechtsen 2018) to run a principal component analysis (PCA) and characterize the genetic structure among subspecies and species. The data were plotted following a custom python script (https://plot.ly/python/v3/ipython-notebooks/principal-component-analysis/). Second, we used a clustering identification approach to estimate admixture proportions across individuals. We ran NGSadmix (Skotte et al. 2013) 15 times for K-values ranging from 1 to 10 and with the remaining settings left at their default. The optimal K-value was determined based on the DeltaK statistic (Evanno et al. 2005), which we calculated from the obtained log-likelihood scores using CLUMPAK (Kopelman et al. 2015). In order to ensure that any potential substructure within continental taxa is detected, we ran independent population-structure analyses once for all individuals and once including only O. mellianus, O. t. traillii, and O. t. robinsoni. Finally, unbalanced allele-sharing in a four-taxon set [((P1,P2),P3),O] can be a sign of post-speciation gene flow (ABBA–BABA test/Patterson’s D; Durand et al. 2011). Building further on the inferred species-tree and cluster identification, we calculated Patterson’s D using doAbbababa2 (Soraggi et al. 2017) to evaluate several putative hybridization scenarios within continental taxa. While O. t. nigellicauda is a visitor to Vietnam (Mason and Allsop 2009), O. t. ardens is not known to migrate to continental territory and geographically more isolated. Therefore, we chose O. t. ardens as outgroup (O) in all comparisons. Then, we computed D-statistics for all possible individual-level comparisons (including one individual per group) and the only possible population-level comparison (including multiple individuals per group). For the individual-based tests, P1 and P2 always consisted of samples belonging to the same (sub-)species, and P3 consisted of a sample belonging to a different taxon as shown in Fig. S1. For the population-level comparison, we used the same software to test for gene-flow between continental taxa. In other words, we checked whether the whole population of O. t. robinsoni (P3), shares more similarities with O. mellianus (P1) than it does with the O. t. traillii (P2) population as shown in Fig. S2.
Genome-wide differentiation between closely related distinct color morphs
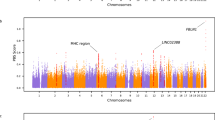
All of the analyses indicated that O. traillii is a paraphyletic species group and that O. t. traillii is likely more closely related to the distinct morph, O. mellianus, than to all other O. traillii populations (Figs. 1c, 2, S3-6). Notwithstanding close phylogenetic association, O. t. traillii and O. mellianus strikingly differ in plumage coloration (Fig. 1b) and we therefore characterized the genetic landscape of differentiation (Fst) to identify candidate loci underlying plumage variation. We measured differentiation by calculating mean Fst (Weir and Cockerham 1984) across nonoverlapping 50 kb windows using VCFtools (V0.1.15) and then visualized the obtained statistics by generating a Manhattan plot using the qqman package (V0.1.4; Turner 2014) in R (V 3.5.0; R Core Team 2018; Fig. 4). Then, we arbitrarily defined outliers as windows with Z(Fst) values over 1.15 (the top 5%) and annotated these by blasting the window sequences to the chicken genome (Gallus.gallus.5.0.cds) using BLAST + (v2.6.0; Camacho et al. 2009). The genes within the top 5% selected windows were then compared with genes involved in plumage coloration in birds. These represent candidate gene regions under selection and thus of potential importance for the development of the distinct differences in plumage between O. t. traillii and O. mellianus.
a Admixture panels showing the best-scoring cluster configurations out of 15 runs produced by NGSadmix for K-values ranging from two to 10. The X axis represents the admixture proportion and the Y axis indicates the sample ID and taxon correspondance. Thick black horizontal lines draw the taxon boundaries according to current classification. Taxa are labeled according to the following abbreviations: ARD: O. t. ardens, NIG: O. t. nigellicauda, ROB: O. t. robinsoni, TRA: O. t. traillii, MEL: O. mellianus. Optimal number of discrete groups returned by DeltaK is 4. B and C: PCA plots showing how samples cluster along continuous axes of variation when including all samples (b) and after excluding island taxa (O. t. ardens and O. t. nigellicauda) (c).
Results
Data processing
The resulting 10X chromium assembly included 37,670 contigs, had a scaffold N50 of 8.3Mbp, and a pseudohaploid assembly length of 1102Mbp. This value is slightly below the genome size of the closely related species O. chinensis, which spans 1398Mbp (C-value = 1.43 pg; Krishan et al. 2005). Out of 303 BUSCOS, 185 (61.1%) were complete and single copy, 2 (0.7%) complete and duplicated, and 116 (38.2%) fragmented or missing. On average, each sample had 267 million raw reads, but this number dropped to 133 million after cleaning, mapping, and excluding short scaffolds (Table S1). The decrease in read numbers was largely caused by the merging of overlapping read pairs. This is expected, given the short DNA templates obtained from historical specimens and does not necessarily reflect loss of sequence information. Furthermore, we found excess of cytosine-to-thymine (C-to-T) misincorporations at 5′-termini and an excess of guanine-to-adenine (G-to-A) misincorporations at 3′-termini (Table S1). After variant calling and filtering, we retained 7.16 million SNPs with GLs and 4.12 million hard-called genotypes.
Phylogenetic framework and divergence-time estimation
Our phylogenetic results unanimously show that O. mellianus forms a well-supported monophyletic lineage that is nested within O. traillii and that O. t. ardens and O. t. nigellicauda (which we will refer to as island subspecies in this paper) form a sister clade that is distantly related to the continental taxa (Figs. 1c, S3–4). Furthermore, the concatenated and full-coalescent phylogeny, as well as the population-structure analyses suggest that O. mellianus is most closely related to O. t. traillii (Figs. 1c, 2, S3). Moreover, this sister relationship is also supported by the mitogenome tree, even though this phylogeny renders O. t. traillii paraphyletic with largely unresolved relationships (Fig. S4). In slight contrast to these results, which collectively suggest that O. mellianus is most closely related to O. t. traillii, the Twisst analyses reveal substantial heterogeneity among nuclear gene trees with three topologies being almost equally frequent (Fig. S7). While O. mellianus is still placed within O. traillii, the relationship between O. mellianus, O. t. traillii, and O. t. robinsoni is therefore less certain.
Based on the nuclear concatenated dataset and the 4.6*E-9 mutation rate of flycatchers by Smeds et al. (2016), the average number of generations since the split between O. mellianus and O. t. traillii was estimated to around 127,000 generations ago (Table S2). Furthermore, the split between these two taxa and O. t. robinsoni was estimated to 224,000 generations ago. Notably, the divergence between O. mellianus and O. t. traillii is less than 2.5 times the estimated number of generations between the most divergent individuals within O. mellianus (ca 52,000 generations) and O. t. traillii (ca 59,000 generations), respectively. As the generation time for O. mellianus and O. traillii is unknown, we cannot estimate the absolute divergence time between major lineages within O. traillii. However, BirdLife International has estimated the generation length for all orioles to 3.7 years, which suggests that the split between O. mellianus and O. t. traillii is younger than 500,000 years.
Introgression analyses
When including all individuals, structure analyses support an optimal number of four clusters, with O. mellianus, O. t. traillii, and O. t. robinsoni forming three and island subspecies grouped into a single cluster (Figs. 2a, b, S6). Similarly, when excluding island taxa, the population-structure analyses support an optimal number of three clusters (Figs. 2c, S5). In both cases, the cluster assignment displayed by the NGSadmix plots is congruent with the PCAngsd analyses (Fig. 2). Furthermore, the order in which the clusters split as the K-value increases and the relative distance between clusters in the PCA plot mirrors the phylogenetic relationships recovered by the nuclear phylogeny (Figs. 1c, 2). For example, O. t. ardens and O. t. nigellicauda, which were found to be most divergent, split off first in the admixture panels and are most distant in the PCA plot (Figs. 1c, 2a, b). O. t. traillii and O. mellianus, which were found to be more closely related, only separate at higher K-values and are adjacent in the PCA plot (Figs. 1c, 2a, b). Across all runs, NGSadmix clearly assigned each individual to its respective taxon and the PCA plot shows very tight clustering of individuals by taxon (Fig. 2b, c, S5-6), indicating that none of the individuals included in this study are recent hybrids.
Positive D-statistics indicate introgression between P2 and P3, while negative D-statistics hint at introgression between P1 and P3. Although the D-statistics were significant for the population, as well as the individual-level comparisons, they tend to be relatively low (<0.03, Fig. 3). The results from the population-based test showed that O. t. traillii and O. t. robinsoni share significantly more site patterns than O. mellianus and O. t. robinsoni (D = 0.0056; Z-score = 3.39; Fig. 3g, Table S3). Furthermore, individual-based tests showed that specimens from the same taxon present different levels of shared derived alleles with individuals from other taxa (Fig. 3a–f, Table S3). D-statistics differed substantially, depending on individuals used (within the same species), suggesting that local populations may have differed in the timing and/or extent of hybridization. For example, when comparing O. t. traillii individuals to each other in order to assess if some are more or less similar to O. t. robinsoni (Fig. 3d), the D-statistics ranged from 0 to 0.015 and O. t. traillii individual Thailand_N7667 tended to be more similar. Moreover, when comparing O. t. traillii individuals to each other in order to contrast levels of similarity with O. mellianus (Fig. 3f), the D-statistics ranged from 0 to 0.023 and O. t. traillii individual India_B9104 tended to be less similar. While the significant excess of shared derived alleles suggests that interspecific gene flow has taken place after initial divergence, relatively low levels of genealogical discordance, clear genetic distinctiveness, and lack of contemporary admixture indicate that gene flow has ceased among contemporary taxa.
The higher the D-statistic, the higher the excess of shared derived alleles between P2 and P3. a–f show the results from the individual-based test. Here, samples were tested individually and shuffled to compute all possible comparisons. g shows the obtained D-statistics from a single population-based comparison in which individuals were grouped by taxon and compared collectively. The significance of the tests is visualized using differently shaped dots. Round dots indicate that the comparison was insignificant (Z-score < 3), while squares indicate that the comparison was significant (Z-score > 3). Individuals discussed in the main text are highlighted according to the taxon-specific color-coding employed throughout the paper. Note that in d, we highlighted the comparisons where Thailand_N7667 was set as P2. On the other hand, in f, we highlighted the comparisons where India_B9104 was set as P1. The outgroup (O) was formed by O. t. ardens but was omitted in this figure for simplicity.
Candidate loci for plumage coloration
The Fst outlier analysis yielded a total of 402 candidate genes when using a 95% cutoff (Z(Fst)>1.15). Among the top 5% outliers were RPE65, ADCY7, CTCF, FCD9, WNT7A, CREB3L1, PRKCB, BNC2, and NCOA6 (Fig. 4). RPE65 is a paralog of BCO2, which is involved in carotenoid degradation to colorless derivatives in white-skinned birds (Toews et al. 2016). CREB3L1, PRKCB, BNC2, and NCOA6 are linked to pigmentation. ADCY7, CTCF, WNT7A, CREB3L1, FCD9, and PRKCB were all differentially expressed between sexes in four plumage patches in zebra finches (Sly 2019). Perhaps most relevant for O. mellianus is CREB3L1, which showed significant signs of differential expression in orange and gray patches in zebra finches. BNC2 and NCOA6 respectively influence skin pigmentation (Jacobs et al. 2013) and hair coloration in humans (Bonilla et al. 2020).
Discussion
Accurately identifying evolutionary units is crucial for conservation and NHC offers a unique opportunity to study endangered taxa. In this study, we assembled a de novo genome, used it as an anchor to guide whole-genome resequencing of museum specimens, and re-evaluated the evolutionary and taxonomic status of O. mellianus and allied oriole species. Our results support that the endangered O. mellianus represents a distinct, recently emerged, and likely reproductively isolated lineage, which merits treatment as a species (De Queiroz 1999). Simultaneously, our results highlight the need for a taxonomic revision of O. traillii ssp., which appears to be a complex comprising several distinct species. We find strong support for a scenario in which O. mellianus has recently emerged from a subspecies of O. traillii and rapidly diversified in plumage coloration. Due to their close affinity, we were able to identify several candidate loci that underlie plumage differentiation and therefore establish a promising research system for the study of speciation.
The systematic affinity of O. mellianus and masked diversity within O. traillii
In line with the results by Jønsson et al. (2019), our analyses show that O. mellianus is nested within O. traillii and consequently O. traillii represents a paraphyletic species complex. Our results consistently recovered the distinct island populations as sister taxon to the continental clade and indicate that O. mellianus is most likely sister to O. t. traillii. While the concatenation and the species-tree approach strongly support a sister relationship between O. mellianus and O. t. traillii (Figs. 1c, S3), the underlying distribution of gene-tree frequencies suggests that a more detailed evaluation is warranted. Topology weighting in the Twisst analysis found high levels of gene-tree discordance with almost equal support for three competing topologies (Fig. S7). The concatenated and Twisst analyses were based on the same dataset, but it is now well established that concatenation masks the underlying heterogeneity in coalescent patterns (Mendes and Hahn 2018). Interestingly, in contrast to concatenation approaches, SNAPP accounts for ILS but also strongly supported an O. mellianus and O. t. traillii sister relationship (Fig. S3). We therefore hypothesize that O. mellianus and O. t. traillii are indeed sisters but diverged shortly after their common ancestor split from O. t. robinsoni. Rapid diversification could lead to a high incidence of ILS and consequently, extensive gene-tree heterogeneity. The discordance among gene-trees was possibly also reinforced by post-divergence gene-flow (see below).
In comparison with previous research (Jønsson et al. 2019), our study includes a large number of O. mellianus and O. traillii representatives, which allows a detailed view on evolutionary relationships at an intraspecific level. By increasing the number of individuals per taxon, we were able to determine that the currently recognized species O. mellianus and the different subspecies of O. traillii (when represented by more than one specimen) form highly supported monophyletic groups according to the nuclear data (O. mellianus and O. t. robinsoni are also monophyletic according to the mitochondrial data; Fig. S4). This is concordant with our PCA and NGSadmix results that recover each nuclear monophyletic lineage as a separate cluster (Fig. 2). Our sampling for O. t. ardens and O. t. nigellicauda is relatively low and this heterogeneity in sample size can introduce biases when filtering variants (Puechmaille 2016). However, repeating population-structure analyses with a different MAF cutoff (0.05) did not change the result, suggesting that island taxa indeed form a monophyletic clade. The high bootstrap support of each clade in the nuclear concatenated tree (Fig. 1c) and the assignment of each individual to its respective cluster (Fig. 2) indicate that O. mellianus, O. t. traillii, and O. t. robinsoni accumulated a substantial amount of unique substitutions. To summarize, our exhaustive dataset confirms that these taxa are genetically cohesive and well-differentiated lineages, suggesting that they evolved independently for a considerable period of time.
Although the mitochondrial phylogeny largely mirrors the branching patterns recovered by analyses based on nuclear data (Figs. 1c, S3–4), we found instances of mitonuclear discordance. The mitogenomic data place O. mellianus as nested within, rather than sister to O. t. traillii and one O. t. traillii specimen (India_B9104) groups with the O. t. robinsoni clade (Fig. S4). This paraphyly could potentially be attributable to ILS or mitochondrial introgression, and differentiating between these two scenarios remains a challenging endeavor (Holder et al. 2001; Buckley et al. 2006 but see Andersen et al. 2020). If ILS is responsible for the observed discordance, it would require retention of shared mitochondrial haplotypes between O. t. traillii and O. t. robinsoni. Twisst points at a relatively rapid radiation of continental taxa. Such radiations are particularly prone to present high levels of ILS. Furthermore, ILS is most likely observed in taxa with large effective population sizes (Maddison 1997; Degnan and Rosenberg 2009). In this respect, it is noteworthy that O. t. traillii has the broadest distribution of all taxa in this study and most certainly the largest population size (Del Hoyo et al. 2008). Mitochondrial introgression, on the other hand, can lead to mitonuclear discordance by horizontally transferring the mitogenome from one taxon into the nuclear background of another. The Indian sample (India_B9104) could hypothetically be carrying an introgressed mitogenome that was donated by O. t. robinsoni after O. t. traillii and O. t. robinsoni diverged. Interestingly, India_B9104 shares the least number of derived alleles with O. mellianus and O. t. robinsoni compared with its conspecifics (Fig. 3f, Table S3). This observation contradicts the hypothesis of mitochondrial introgression, though mitochondrial introgression has previously been reported with negligible signs of nuclear introgression (Good et al. 2015; Andersen et al. 2020). Taken together, the mitonuclear discordance can be explained by both post-divergence gene flow and ILS. While we have detected biased excess of discordant genealogies stemming from post-divergence gene flow, we cannot discard that ILS is also leading to random gene-tree discordance. ILS and introgression are non-mutually exclusive evolutionary processes and can act together, leading to discordant coalescent histories.
Evidence for gene-flow
Building further on our population-level sampling for several (sub-) species, we find no evidence for contemporary gene flow among lineages. However, explicit tests for introgression reported significant D-statistics (Z-score > 3; Fig. 3, Table S3). While the observed D-statistics are relatively low (D < 0.03), the ABBA–BABA analyses are parsimonious in terms of geography. For instance, the population-level comparison showed that O. t. traillii and O. t. robinsoni, which have adjacent breeding-distribution ranges, share a significant excess of derived alleles compared with O. mellianus (Figs. 1a, 3g, Table S3). However, this result should be interpreted with caution, given that in this comparison, we have followed the concatenated nuclear phylogeny: [(O. mellianus, O. t. traillii),O.t.robinsoni]. The results of our Twisst analysis equally support the three possible in-group topologies, suggesting that this assumption may be violated (Fig. S7). Even so, individual-based tests also show geographically parsimonious signals. In these comparisons, we assessed if individuals of the same taxon share more or less similarities with another taxon. They are therefore valid under all topologies supported by Twisst. For example, the O. t. traillii sample India_B9104, is geographically most isolated and also has the lowest signs of introgression with O. t. robinsoni and O. mellianus (Figs. 1a, 3f, Table S3). Conversely, Thailand_N7667 is the O. t. traillii sample in closest geographic proximity to O. t. robinsoni and displays the highest signs of introgression with this taxon (Figs. 1a, 3d, Table S3).
The significance and geographic correlation of our results suggest that the observed excess of discordant genealogies may stem from a rapid radiation, followed by post-divergence gene flow between O. t. traillii and O. t. robinsoni. Ancestral hybridization, rather than recent gene flow, could explain why NGSadmix detected no admixture (Fig. 2a). The ABBA–BABA test explicitly focuses on the frequency of non-concordant site patterns and is capable of detecting historical gene flow, while NGSadmix is only sensitive to recent admixture (Skotte 2015). It could be argued that recent gene flow was not detected because of the lack of samples coming from putative hybrid zones, where admixture is more likely. Thus, we recommend that future studies include additional samples, particularly from areas where these taxa meet. However, genome-wide cluster-based analyses and explicit introgression tests performed on the available data did not support a scenario in which O. mellianus is nested within O. traillii due to recent hybridization, as hypothesized by Jønsson et al. (2019). Instead, given the observed genome-wide signal that consistently positions O. mellianus in close relationship to continental O. traillii subspecies, it seems uncontested that they truly share recent common ancestry.
Taxonomic Implications
Our findings suggest that the current taxonomic classification does not align with the evolutionary history of the group. O. traillii is paraphyletic and most of the subspecies within this complex have been reproductively isolated for a long time, certainly longer than the currently recognized species O. mellianus and the subspecies O. t. traillii. In other words, from a phylogenetic perspective, different O. traillii subspecies should be elevated to full species if O. mellianus is treated as a distinct species, which is warranted, given the lack of extant gene flow. We did not intend to revalidate or describe new species in this contribution. However, given the observed inconsistency, we use the population genetic and phylogenetic results provided by whole nuclear and mitochondrial genomes to suggest a tentative taxonomic classification in which O. t. traillii and O. t. robinsoni are elevated to species rank. While we strongly suspect that the two island subspecies of the maroon oriole form reciprocally monophyletic lineages, more data are needed to make that statement and thus, we recommend to provisionally consider them as a single species for which the name O. t. ardens has priority.
We note that our geographic and genetic sampling is exhaustive, but a proper assessment of species limits should draw on genetic as well as ecological, morphological, behavioral, and acoustic data (Tobias et al. 2010). Hence, we do not include a full taxonomic description in this paper, and recommend considering such information in greater detail before formally describing new species within O. traillii. Interestingly, despite being more homogeneous in terms of appearance, O. traillii subspecies do exhibit some geographic variation in plumage tones, size, bill shape, and migratory behavior (Vigors 1832; Swinhoe 1870; Stresemann 1922; Delacour 1927). An integrative taxonomic approach, which our study is one part of, is urgently needed and may lead to a significant reassessment of the conservation status of several (sub-)species (see below).
Color genes under selection
Asian Orioles revealed marked differences in the rate and degree of phenotypic diversification and O. mellianus has developed its unique plumage coloration relatively recently. This observation raises the question whether color differentiation promoted by sexual selection has driven the divergence of O. mellianus. Plumage coloration, particularly in exposed patches such as the breast and the mantle, plays important roles in social signaling and mate choice in many birds (Hill and McGraw 2006a, 2006b). O. traillii males are coated with different tones of crimson-maroon depending on the subspecies and O. mellianus males are coated silvery white whereas females of both taxa resemble a somewhat duller version of the males (Del Hoyo et al. 2008). Therefore, plumage coloration is probably subject to female preference. One can thus speculate that color-assortative mate choice was involved in the initial separation of O. mellianus.
By comparing the genome-wide landscape of differentiation between O. mellianus and O. t. traillii, we identified strong candidate genes that may be important for the development of the distinct plumage of O. mellianus. Several of them were already previously found to be involved in avian plumage coloration (Fig. 4). However, determining how a genotype generates particular plumage phenotypes is difficult. Confounding factors such as demographic history and heterogeneous mutation rates, recombination rates, and background selection across the genome can obscure real signature selection or create false-positive signals (Ravinet et al. 2017). Furthermore, plumage-color difference in this omnivorous-species group can also be attributable to variation in dietary preference (LaFountain et al. 2013) or regulated by an interaction between multiple genes (Roulin and Ducrest 2013). A detailed understanding of the genetic basis behind the plumage differences is beyond the scope of the present study, but the identification of several candidate loci that match plumage differentiation in other avian systems is promising (Poelstra et al. 2014; Toews et al. 2016; Campagna et al. 2017). We hope that our results stimulate further attempts to understand the drivers of speciation among orioles and that they serve as a starting point for more thorough studies on the evolution of the marked color differences between O. mellianus and O. traillii.
Implications for conservation
With the recent update of China’s endangered-species list, O. mellianus is now legally protected on a national level in China and Thailand as it is threatened by habitat loss and continuing declines appear to be occurring in most regions. Unfortunately, to our knowledge, no active conservation-management program is specifically targeting this species. Nonetheless, there is an urgent need to design a systematic monitoring scheme to clarify habitat and conservation requirements in order to design a suitable recovery plan (BirdLife International 2016). By outlining the genetic uniqueness and species status of this taxon, we hope to encourage authorities to intensify the protection of suitable habitat in order to preserve the remaining O. mellianus populations. Furthermore, we hope to promote further research focused on obtaining crucial knowledge for conservation, such as the distribution, demography, life history, and ecology of this species.
Moreover, the endemic O. t. ardens is currently estimated to have a declining population size of 200–500 breeding pairs (Fang 2005) and is considered endangered by the Taiwan Wildlife Conservation Act. We hope that our findings highlight the urgency for conservation prioritization since they, together with O. t. nigellicauda, present a deeply divergent lineage. As most conservation programs still rely on taxonomic (species) designations and our study revealed that the taxonomy currently underestimates the lineage diversity of O. traillii subspecies, it is urgent to re-evaluate current conservation schemes so that prioritization is not only proportional to phenotypic differences, but also considers genetic variation. Furthermore, we hope that our contribution encourages further research to study the patterns of genetic differentiation between O. t. ardens and O. t. nigellicauda since this could have important implications for the conservation of the Taiwanese maroon oriole subspecies O. t ardens.
Conclusion
Understanding the evolutionary history and processes that have shaped extant populations is critical for conservation. However, it is often challenging to conduct such studies with sampling being a major hurdle (Thompson 2013). Here, we show how modern genomic technologies and NHC can be combined to reveal hidden diversity, evaluate population connectivity, and how this can ultimately guide conservation considerations. Whereas the integration of historical specimens has been long hampered by a lack of genomic resources, we demonstrate that it is now feasible and cost-effective to generate de novo reference genomes that can guide evolutionary analyses needed for both taxonomic and conservation purposes. Finally, our findings illustrate that an integrative approach is needed to identify taxonomic units for conservation, that species delineation solely based on morphology can underestimate genetic divergence, and that such studies remain vital even in relatively well-known organismal groups such as avian vertebrates.
Data availability
The raw resequenced data, the 10X chromium reads, and the 10X reference assembly have been deposited with Dryad (https://datadryad.org/stash/share/bJR2tGU20m2wZ2mWQ2z1MPik7eBkd6aXlgi3ZX341LA).
References
Andersen MJ, McCullough JM, Gyllenhaal EF, Mapel XM, Haryoko T, Jønsson KA et al. (2020) Complex histories of gene flow and a mitochondrial capture event in a non‐sister pair of birds. Mol Ecol 30:2087–2103
Andrews S (2010) Fastqc: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Astrin JJ, Höfer H, Spelda J, Holstein J, Bayer S, Hendrich L et al. (2016) Towards a DNA barcode reference database for spiders and harvestmen of Germany. PloS ONE 9:e0162624
Bi K, Linderoth T, Vanderpool D, Good JM, Nielsen R, Moritz C (2013) Unlocking the vault: next‐generation museum population genomics. Mol Ecol 22:6018–6032
BirdLife International (2016) Oriolus mellianus. IUCN Red List Threatened Species 2016:e.T22706449A94070575
BirdLife International (2018) Oriolus traillii. IUCN Red List Threatened Species 2018:e.T22706446A130378420
Blom MPK (2021) Opportunities and challenges for high‐quality biodiversity tissue archives in the age of long‐read sequencing. Mol Ecol 30:5935–5948
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Bonilla C, Bertoni B, Min JL, Hemani G, Genetics of DNA Methylation Consortium, Elliott HR (2020) Investigating DNA methylation as a potential mediator between pigmentation genes, pigmentary traits and skin cancer. Pigment Cell Melanoma Res 00:1–13
Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A (2012) Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol Biol Evol 29:1917–1932
Buckley TR, Cordeiro M, Marshall DC, Simon C (2006) Differentiating between hypotheses of lineage sorting and introgression in New Zealand alpine cicadas (Maoricicada Dugdale). Syst Biol 55:411–425
Burrell AS, Disotell TR, Bergey CM (2015) The use of museum specimens with high-throughput DNA sequencers. J Hum Evol 79:35–44
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009) BLAST+: architecture and applications. BMC Bioinforma 10:1–9
Campagna L, Repenning M, Silveira LF, Fontana CS, Tubaro PL, Lovette IJ (2017) Repeated divergent selection on pigmentation genes in a rapid finch radiation. Sci Adv 3:e1602404
Christmas MJ, Jones JC, Olsson A, Wallerman O, Bunikis I, Kierczak M et al. (2021) Genetic barriers to historical gene flow between cryptic species of alpine bumblebees revealed by comparative population genomics. Mol Biol Evol 38:3126–3143
Costello MJ, Beard KH, Corlett RT, Cumming GS, Devictor V, Loyola R et al. (2016) Field work ethics in biological research. Biol Conserv 203:268–271
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Degnan JH, Rosenberg NA (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol 24:332–340
Delacour JT (1927) Oriolus traillii robinsoni. Bull Br Ornithol Club 47:168–169
Del Hoyo J, Elliot A, Christie DA (2008) Handbook of the birds of the world Vol 13: Penduline tits to Shrikes. Lynx Edicions, Barcelona
De Queiroz K (1999) The general lineage concept of species and the defining properties of the species category. In: Wilson RA (ed.) Species: New Interdisciplinary Essays: 49–89
Durand EY, Patterson N, Reich D, Slatkin M (2011) Testing for ancient admixture between closely related populations. Mol Biol Evol 28:2239–2252
Dussex N, Taylor HR, Irestedt M, Robertson BC (2018) When genetic and phenotypic data do not agree: the conservation implications of ignoring inconvenient taxonomic evidence. N Z J Ecol 42:284–290
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewels P, Magnusson M, Lundin S, Käller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048
Fang WH (2005) A guide to threatened birds of Taiwan. Owl Publishing House, Taipei
Garrison E (2012) Vcflib: A C++ library for parsing and manipulating VCF files. https://github.com/ekg/Vcflib
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/abs/1207.3907
Good JM, Vanderpool D, Keeble S, Bi K (2015) Negligible nuclear introgression despite complete mitochondrial capture between two species of chipmunks. Evolution 69:1961–1972
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075
Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - A baiting and iterative mapping approach. Nucleic Acids Res 41:e129
Hill GE, McGraw MJ (2006a) Bird coloration volume I: mechanisms and measurement. Harvard University Press, Cambridge
Hill GE, McGraw MJ (2006b) Bird coloration volume II: function and evolution. Harvard University Press, Cambridge
Holder MT, Anderson JA, Holloway AK (2001) Difficulties in detecting hybridization. Syst Biol 50:978–982
Holmes MW, Hammond TT, Wogan GOU, Walsh RE, LaBarbera K, Wommack EA et al. (2016) Natural history collections as windows on evolutionary processes. Mol Ecol 25:864–881
Howell N, Elson JL, Howell C, Turnbull DM (2007) Relative rates of evolution in the coding and control regions of African mtDNAs. Mol Biol Evol 24:2213–2221
Irestedt M, Ohlson JI, Zuccon D, Källersjö M, Ericson PGP (2006) Nuclear DNA from old collections of avian study skins reveals the evolutionary history of the Old World suboscines (Aves, Passeriformes). Zool Scr 35:567–580
Jacobs LC, Wollstein A, Lao O, Hofman A, Klaver CC, Uitterlinden AG et al. (2013) Comprehensive candidate gene study highlights UGT1A and BNC2 as new genes determining continuous skin color variation in Europeans. Hum Genet 132:147–58
Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L (2013) mapDamage2. 0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29:1682–1684
Jønsson K, Blom MPK, Marki PZ, Joseph L, Sangster G, Ericson PGP et al. (2019) Complete subspecies-level phylogeny of the Oriolidae (Aves: Passeriformes): Out of Australasia and return. Mol Phylogenet Evol 137:200–209
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinforma 15:356
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455
Krishan A, Dandekar P, Nathan N, Hamelik R, Miller C, Shaw J (2005) DNA index, genome size, and electronic nuclear volume of vertebrates from the Miami Metro Zoo. Cytom A 65:26–34
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
LaFountain AM, Frank HA, Prum RO (2013) Carotenoids from the crimson and maroon plumages of Old World orioles (Oriolidae). Arch Biochem Biophys 539:126–132
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25:2078–2079
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013)
Maddison WP (1997) Gene trees in species trees. Syst Biol 46:523–536
Martin SH, Van Belleghem SM (2017) Exploring evolutionary relationships across the genome using topology weighting. Genetics 206:429–438
Mason P, Allsop J (2009) The golden oriole. A&C Black, London
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
McMahon BJ, Teeling EC, Höglund J (2014) How and why should we implement genomics into conservation? Evolut Appl 7:999–1007
Meisner J, Albrechtsen A (2018) Inferring population structure and admixture proportions in low depth next-generation sequencing data. Genetics 210:719–731
Mendes FK, Hahn MW (2018) Why concatenation fails near the anomaly zone. Syst Biol 67:158–169
Meyer M, Kircher M (2010) Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc 6:pdb-prot5448
Miller CR, Waits LP (2003) The history of effective population size and genetic diversity in the Yellowstone grizzly (Ursus arctos): implications for conservation. PNAS 100:4334–4339
Mondol S, Bruford MW, Ramakrishnan U (2013) Demographic loss, genetic structure and the conservation implications for Indian tigers. Proc R Soc B 280:20130496
Ortiz EM (2019) Vcf2phylip v20: Convert a VCF matrix into several matrix formats for phylogenetic analysis. https://github.com/edgardomortiz/vcf2phylip
Payne RB, Sorenson MD (2002) Museum collections as sources of genetic data. Bonn Zool Bull 51:97–104
Petersen KR, Streett DA, Gerritsen AT, Hunter SS, Settles ML (2015) Super Deduper, fast PCR duplicate detection in fastq files. Proc 10th ACM Int Conf Bioinform Comput Biol Health Inform: 491–492
Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, Müller I et al. (2014) The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science 344:1410–1414
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 17. Syst Biol 67:901–904
Raven PH, Miller SE (2020) Here today, gone tomorrow. Science 370:149–149
Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlović M et al. (2017) Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J Evol Biol 30:1450–1477
R Core Team (2018) R: A language and environment for statistical computing. https://www.R-project.org/
Rheindt FE, Szekely T, Edwards SV, Lee PL, Burke T, Kennerley PR et al. (2011) Conflict between genetic and phenotypic differentiation: the evolutionary history of a ‘lost and rediscovered’ shorebird. PloS ONE 6:e26995
Roulin A, Ducrest AL (2013) Genetics of coloration in birds. Sem Cell Dev Biol 24:594–608
Schloss JA, Gibbs RA, Makhijani VB, Marziali A (2020) Cultivating DNA sequencing technology after the human genome project. Annu Rev Genomics Hum Genet 21:117–138
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Skotte L, Korneliussen TS, Albrechtsen A (2013) Estimating individual admixture proportions from next generation sequencing data. Genetics 195:693–702
Skotte L (2015) Statistical Approaches Accomodating Uncertainty in Modern Genomic Data. Dissertation, University of Copenhagen
Sly ND (2019) The genetic mechanisms underlying pigmentation and their evolutionary importance in birds. Dissertation, University of Montana
Smeds L, Qvarnström A, Ellegren H (2016) Direct estimate of the rate of germline mutation in a bird. Genome Res 26:1211–1218
Soraggi S, Wiuf C, Albrechtsen A (2017) Powerful inference with the D-statistic on low-coverage whole-genome data. G3 8: 551-566Stamatakis A (2014) RAxML Version 8: A tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics 30:1312–1313
Stanton DW, Frandsen P, Waples RK, Heller R, Russo IRM, Orozco-terWengel PA et al. (2019) More grist for the mill? Species delimitation in the genomic era and its implications for conservation. Conserv 20:101–113
Stresemann E (1922) Oriolus traillii mellianus n subsp. Ornitol Mon Ber 30:64
Supple MA, Shapiro B (2018) Conservation of biodiversity in the genomics era. Genome Biol 19:1–12
Swinhoe R (1870) On the ornithology of Hainan. Ibis 6:363–365
Thompson W (2013) Sampling rare or elusive species: concepts, designs, and techniques for estimating population parameters. Island Press, Washington
Tobias JA, Seddon N, Spottiswoode CN, Pilgrim JD, Fishpool LDC, Collar NJ (2010) Quantitative criteria for species delimitation. Ibis 152:724–746
Toews DPL, Taylor SA, Vallender R, Brelsford A, Butcher BG, Messer PW et al. (2016) Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr Biol 17:2313–2318
Turner SD (2014) qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Preprint at https://doi.org/10.1101/005165v1.full
Vigors NA (1832) Pastor traillii. Proc Zool Soc Lond 1831:175
Walters MJ (2006) Do no harm. Conserv Pract 7:28–34
Wandeler P, Hoeck PE, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends Ecol Evol 22:634–642
Weir BS, Cockerham CC (1984) Estimating F-Statistics for the analysis of population structure. Evolution 38:1358–1370
Winker K, Reed JM, Escalante P, Askins RA, Cicero C, Hough GE et al. (2010) The importance, effects and ethics of bird collecting. Auk 127:690–695
Zhang J, Kobert K, Flouri T, Stamatakis A (2014a) PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620
Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C et al. (2014b) Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgements
Footpad samples for genetic analyses were generously provided by American Museum of Natural History, New York (Paul Sweet, Tom Trombone, and Peter Capainolo), British Museum of Natural History (Hein Van Grouw and Mark Adams), Smithsonian National Museum of Natural History (Christopher M. Milensky), Natural History Museum of Denmark (Jan Bolding), and Swedish Museum of Natural History, Stockholm, Sweden (Ulf Johansson). MI acknowledges support from the Swedish Research Council (grant Nos. 2019-03900 and 621-2014-5113). The authors acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. Moreover, the authors extend special gratitude to Carolina Diaz Roman for illustrating the bird figures.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
MI, MB, KAJ, and ME conceived the study. MI procured the samples and carried out the lab work. ME performed the bioinformatic- and data analyses with supervision from MB, except the genome scan, which was performed by PE and MI. ME, MI and MB interpreted the results. ME wrote the paper with input from all co-authors.
Corresponding author
Ethics declarations
COMPETING INTERESTS
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Pår Ingvarsson.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ernst, M., Jønsson, K.A., Ericson, P.G.P. et al. Utilizing museomics to trace the complex history and species boundaries in an avian-study system of conservation concern. Heredity 128, 159–168 (2022). https://doi.org/10.1038/s41437-022-00499-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-022-00499-0