Abstract
Background:
Mild brain hypothermia (31–34 °C) after neonatal hypoxia–ischemia (HI) improves neurodevelopmental outcomes in human and animal neonates. Using an asphyxia model with neonatal mice treated with mild hypothermia after HI, we investigated whether 1H nuclear magnetic resonance (NMR) metabolomics of brain extracts could suggest biomarkers and distinguish different treatments and outcome groups.
Methods:
At postnatal day 7 (P7), CD1 mice underwent right carotid artery occlusion, 30 min of HI (8% oxygen), and 3.5 h of either hypothermia (31 °C) or normothermia (37 °C). Whole brains were frozen immediately after HI, immediately after 3.5 h of hypothermia or normothermia treatments, and 24 h later. Perchloric acid extractions of 36 metabolites were quantified by 900 MHz 1H NMR spectroscopy. Multivariate analyses included principal component analyses (PCA) and a novel regression algorithm. Histological injury was quantified after HI at 5 d.
Results:
PCA scores plots separated normothermia/HI animals from hypothermia/HI and control animals, but more data are required for multivariate models to be predictive. Loadings plots identified 11 significant metabolites, whereas the regression algorithm identified 6. Histological injury scores were significantly reduced by hypothermia.
Conclusion:
Different treatment and outcome groups are identifiable by 1H NMR metabolomics in a neonatal mouse model of mild hypothermia treatment of HI.
Similar content being viewed by others
Main
Although mild therapeutic hypothermia after neonatal asphyxia improves outcomes and is now a standard of care, further insight into patient management is needed from studies on brain injury and recovery mechanisms, as well as into challenges in predicting outcomes. The ability to modify genes and their expression has made mouse animal models useful in elucidating pathological mechanisms. Publications already exist of murine investigations of hypothermia as a treatment for hypoxic–ischemic (HI) brain injury (1,2,3,4). Various murine-based investigations of other pathologies have found 1H nuclear magnetic resonance (NMR)–based metabolomics to be useful (5,6,7). In a recent oxygen–glucose deprivation study with superfused respiring neonatal rat brain slices, we used 1H NMR–based metabolomics of perchloric acid brain extracts to distinguish recovery groups belonging to different post–oxygen–glucose deprivation temperature treatments and to identify responsible metabolites (8). Large sample sizes, common in metabolomics studies of biofluids that can be obtained relatively easily, such as blood and urine, are often impractical and too costly in preclinical laboratory neuroscience studies. However, studies with smaller sample sizes can be useful if they produce experimental differences that are significantly large. In this study, 1H NMR at 21.1 Tesla (900 MHz) was used for metabolomic analyses of whole brains, obtained from neonatal mice in an in vivo model of neonatal HI. If brain metabolomics can distinguish normothermic from hypothermic posttreatment data sets, future studies that add outcome data can address whether brain metabolomics can help guide therapy, predict outcomes, and become clinically useful.
Results
Hypothermia Reduced Histological Injury 5 d After HI
Figure 1 shows that mild hypothermia reduced brain injury in our model. The median injury score was 22.5 (range: 3–24) for the normothermia group as compared with 6 (range 3–20) for the mild hypothermia group. Therefore, mice that received hypothermia had lower injury scores as compared with mice treated with normothermia (P = 0.005). The overlap of the distributions is small enough to consider using the terms “strong injury” and “mild injury,” using 14 as the dividing score. Doing so leads to the conclusion that the hypothermia group had 2.0 ± 1.3 mice (17 ± 11%) with strong injury, as compared with 9.0 ± 1.5 mice (75 ± 13%) with strong injury in the normothermia group. A two-sample binomial proportion test to determine if there is a statistical difference between 9/12 and 2/12 resulted in a P value of 0.014.
Histological injury scores of mouse brains obtained after 5 d of recovery from hypoxia–ischemia (HI). Data at the left in red show scores for 12 mice kept normothermic after HI, whereas the data at the right in blue provide scores for 12 mice that were treated after HI with 3.5 h of mild hypothermia (31 °C). Median injury scores are shown by black horizontal bars. Mice that received hypothermia had lower injury scores as compared with mice treated with normothermia (P = 0.005).
Nine Metabolites Were Identified in Pairwise t-Tests
Figure 2 uses three vertical bar graphs to compare the quantifications of all 36 brain metabolites in five groups: the control group (control), the postnatal day 7 (P7) hypothermia group (P7 hypothermia), the P7 normothermia group (P7 normothermia), the P8 hypothermia group (P8 hypothermia), and the P8 normothermia group (P8 normothermia). The detailed descriptions for each experimental group are provided in the Methods section. Figure 2a shows metabolites that had very significant metabolite changes, whereas Figure 2b shows metabolites to the left of the 4.67 parts-per-million (ppm) water NMR peak, where metabolite signals are not detectable in vivo but are distinguishable in this study because of the high magnetic field (21.1 Tesla) and high spectral resolution (~0.002 ppm). Figure 2c shows the remaining metabolites. Eight of the nine metabolites identified in the t-tests were in Figure 2a : glutamine, acetate, succinate, aspartate, formate, N-acetyl-aspartyl-glutamate, isoleucine, and taurine. The ninth metabolite, fumarate, is in Figure 2b . Comparison results are summarized in Tables 1 and 2 .
Vertical bar graphs of all 1H nuclear magnetic resonance (NMR) metabolites. Markings relate to results of pairwise comparison testing between groups: control vs. P7 hypothermia; control vs. P7 normothermia; control vs. P8 hypothermia; control vs. P8 normothermia; P7 hypothermia vs. P7 normothermia; and P8 hypothermia vs. P8 normothermia. The five vertical bars with SEs for each metabolite correspond to protocol times when animal brains were taken. From left to right, the first bar refers to control animals (no hypoxia–ischemia (HI)), whereas the remaining four bars are for animals that underwent 30 min of HI followed immediately by 1 h of normoxia with the dam, followed by 3.5 h of normothermia or mild hypothermia, and then either no further recovery or 24 h of recovery. The vertical axis is for metabolite intensities normalized to dry powder weights and also to the 1H signal intensity to the left of the DSS peak. Below each set of vertical bars is the multiplication factor used to raise each set’s height to a more visible range. (a) Metabolites with statistically significant differences in between-treatment groups (*P < 0.05, †P < 0.01). (b) Metabolites with NMR peaks to the left of the water peak in Figure 1c (*P < 0.05). (c) Metabolite quantifications not included in graphs a and b. DSS, 4,4-dimethyl-4-silapentane-1-sulfonic acid; GABA, γ-aminobutyric acid; Gpcho, glycerophosphocholine; NAAG, N-acetyl-aspartyl-glutamate; P, postnatal day; PCho, phosphocholine; PCr, phosphocreatine.
Separate Clusters of Hypothermia and Normothermia Groups in Principal Component Analyses
In Figure 3a , the scores plot for the plane of the first two principal component axes (R2 = 0.55; Q2 = 0.25) shows a clear separation of the HI group from all end of recovery groups, which overlap among themselves. In the corresponding loadings plot ( Figure 3b ), highlighting of the HI group’s vector components shows that the HI group’s separation is due to increases in levels of alanine, ADP, choline, lactate, succinate, valine, γ-aminobutyric acid, and isoleucine, and decreases in levels of ATP, phosphocreatine (PCr), phosphocholine (PCho), malate, aspartate, taurine, and N-acetyl-aspartate.
Principal component analysis (PCA) scores and loadings plots for 18 data points, three from the end of HI, and 15 from the end of the recovery period. (a) Scores plot: horizontal and vertical axes are, respectively, for PC1 and PC2. Colors: yellow, control; light green, P7 hypothermia; light blue, P8 hypothermia; purple, P7 normothermia; reddish brown, P8 normothermia; black, HI. In a, black HI data are bunched to the right, separated from the outcome groups. R2 values for PC1 and PC2 are 0.386 and 0.167, respectively. (b) Loading plot: metabolites responsible for separations are shown in black bold print, next to brown dots; metabolites are shown in gray print next to green dots. GABA, γ-aminobutyric acid; Gpcho, glycerophosphocholine; HI, hypoxia–ischemia; NAAG, N-acetyl-aspartyl-glutamate; P, postnatal day; PCho, phosphocholine; PCr, phosphocreatine.
Similarly, the scores plot in Figure 4a (R2 = 0.563; Q2 = 0.002) for the ensemble of control and outcome groups shows a separation of the P8 normothermia group from the other groups, which are comingled. In the corresponding loadings plot ( Figure 4b ), highlighting of the P8 normothermia group’s vector components shows that the metabolite changes most responsible for the cluster separation were increases in glutamine, fumarate, succinate, isoleucine, N-acetyl-aspartyl-glutamate, acetate, and formate, and decreases in taurine, histidine, malate, and ascorbate.
Principal component analysis (PCA) for the five outcome groups compared in Figure 2 . Data were analyzed exactly as in Figure 3 , after excluding the three HI data points. (a) Scores plot: data for the P8 normothermia group (red squares) are clustering in the right lower quadrant. R2 values for PC1 and PC2 are 0.243 and 0.175, respectively. (b) Loadings plot: metabolites contributing to the separation are highlighted in red and blue. (c) Contribution plot between the groups, as described in the text. Vertical green bars are distances in multivariate space between average metabolite vectors for each group. For both b and c, points in red and blue correspond to P8 hypothermia group metabolites that are increased and decreased, respectively. GABA, γ-aminobutyric acid; Gpcho, glycerophosphocholine; HI, hypoxia–ischemia; NAAG, N-acetyl-aspartyl-glutamate; P, postnatal day; PCho, phosphocholine; PCr, phosphocreatine.
Construction of the contribution plot ( Figure 4c ) began by calculating, one metabolite at a time, the average multivariate space vector for each metabolite in the P8 normothermia group. After bunching together all remaining treatment groups, the software similarly calculated each metabolite’s average vector for the bunch. Green bars show, metabolite by metabolite, the magnitudes of vectors obtained by subtracting the average metabolite vectors in the P8 normothermia group from their partners in the bunched group. Numbers are plotted as positive or negative according to whether or not they are decreased or increased in the P8 normothermia group. Colored dots coordinate metabolite locations in Figure 4b with locations in Figure 4c .
Regression Analyses: Good Linear Models Require Fewer Than Four Metabolites
Results of the L1-penalized lasso regression analysis, performed with PCr as the outcome variable, are shown in Figure 5 . The graph in Figure 5a shows the relationship between model predictive performance, as measured via mean squared error (MSE, y-axis), and model complexity, as measured via log(λ) (x-axis). The corresponding number of metabolites for a good linear model is given on the upper x-axis, at the top of the figure. Therefore, the PCr outcome results suggest that three to four variables are all that are needed to provide a best fit.
Plots produced by the L1-penalized lasso regression. (a) The vertical axis is for mean-squared errors (MSEs) from the regression’s cross-validation procedure, plotted as a function of log(λ), shown along lower horizontal axes. λ Represents the tuning parameter. Numbers along the upper horizontal axis represent the average number of predictor variables, n, for linear models for different values of log(λ). Going along a horizontal axis from right to left has λ decreasing toward zero and n increasing toward 35. Vertical black lines bracket n values, where the model provides its best fits to the data. Red dots indicate average MSE values for all models resulting from the corresponding value of λ, and vertical bars through the red dots show upper to lower MSE values. (b) Coefficient trajectories produced by the regression analysis (in a). The lower horizontal axis shows L1, the λ-dependent sum of absolute values of (predicted − observed) differences. The upper horizontal axis indicates the number of predictor values shown in a. When going from left to right, λ is decreasing toward zero. PCho, phosphocholine.
Features of the graph that indicate very good success in the fitting include an initial drop in MSE, going from left to right, as the value of λ starts to increase from ≈0 (10−7.5, because the log value is −7.5). The fits improve until the lowest point in the dip because unimportant metabolites are excluded and important metabolites increasingly show dominance. At a λ value near 10−5.7, the MSE has almost no change with further increases in λ, suggesting that several statistical models obtain approximately equal and minimal MSE. Finally, as λ increases beyond 10−5.2, one has ever-increasing values of MSE because not enough important variables are included. A companion plot ( Figure 5b ) shows coefficient trajectories as successive metabolites enter the model, corresponding to relaxation of the L1 penalty by decreasing λ toward zero, going from left to right. Figure 5b shows that the top six 1H NMR metabolites were ATP-ADP, PCho, lactate, glucose, valine, and glutamate.
Discussion
The most important finding in this study is that although no metabolomics differences were apparent immediately after terminating the mild hypothermia treatment, at 24 h post-HI in the scores plot ( Figure 4a ), an impressive separation can be seen in the P8 normothermia group relative to the hypothermia and control groups, which are indistinguishable from each other.
The very clear separation of HI data sets from all others in the principal component analysis (PCA) scores plot ( Figure 3a ), which identified energy-related metabolites as primarily responsible, is more comforting than surprising, because HI brains were obtained at the time of maximal insult, before normoxia was restored, when conditions are physiologically very different from all other times. Other responsible variables included valine, a branched-chain amino acid that returns nitrogen to astrocytes in neuron–astrocyte glutamate recycling and that is also known to be increased during excitotoxicity (9). Astrocytic uptake of synaptic glutamate is followed by its amidation to glutamine. After glutamine is passed to presynaptic neurons for recycling into glutamate, the amino nitrogen taken from glutamine is returned to astrocytes via branched-chain amino acids (10).
In the HI and P8 normothermia groups, respectively, increased and decreased tricarboxylic acid cycle intermediates succinate and malate can be linked to energy metabolism (11). The malate–aspartate shuttle is the primary mechanism for maintaining correct cytoplasmic and mitochondrial nicotinamide adenine dinucleotide oxidized/nicotinamide adenine dinucleotide reduced ratios, whereas succinate contributes to complex II on the mitochondrial membrane. Lactate, alanine, and citrate can be transferred from astrocytes to neurons for metabolic support. The osmotic regulator taurine and the important neuron marker N-acetyl-aspartate are known to be decreased in neonatal HI.
One report (12) using sensorimotor and water maze tests found persistent neurological deficits at 7 and 30 d after HI. Those results, together with our histology results, suggest that future HI murine studies with both outcome and metabolite data sets are likely to identify the metabolites most responsible for metabolomics differences and to evaluate whether they are early predictors of long-term neurological outcomes.
Both PCA scores plots ( Figures 3a and 4a ) had R2 values >0.5, indicating that the two PCA statistical models explained >50% of each plot’s total variance. However, for both PCA analyses, the Q2 values were <0.25, which is less than the desired minimum value of 0.5. Therefore, although the finding of different clustering in multivariate space is significant, the model created from our data set was not sufficiently robust to be predictive. Clearly, we have too few data points for sustaining a PCA cross-validation procedure that eliminates input data. In a clinical setting, one would need enough statistics to have predictive data sets, because these are used to alter management, especially to try avoiding poor outcomes when they are predicted.
The contribution plot ( Figure 4c ), which presents lengths of vectors in the multidimensional multivariate space, confirms the clustering suggested in Figure 4b , which displays two-dimensional (2D) distances that are qualitative in a visual inspection of Figure 4b . Moreover, it confirms that there are no outliers and that identification of biomarker candidates is the same for both plots.
The L1-penalized regression algorithm was very successful in identifying the metabolites and roles of the most important biomarkers. This method supplemented our PCA approach to biomarker identification, which is most successful in situations in which there are many multivariate data sets (“large N”), and thus enough points on the scores plot for different validation tests of cluster separation. In PCA, the identity of metabolites responsible for separate clusters is found in the loadings plots.
The L1-penalized regression identifies biomarkers in situations for which it is not practical to have a very large N, which is the case in preclinical animal studies such as the current one and others, for which each point in a multivariate data set costs thousands of dollars. For our small-N data set, the L1-penalized regression analysis produced easily recognizable minima in the MSE plots in Figure 5a , b . PCr is an outcome variable associated with a healthy state of high-energy phosphates, and the metabolites found by the algorithm are mechanistically relevant, with roles being apparent in Figure 5b . All of the metabolites with negative coefficients occur in larger quantities when there is more injury and PCr depletion, whereas metabolites with positive coefficients occur in larger quantities when there is PCr preservation or augmentation. PCho increases can be produced by increased PCho synthesis via choline kinase, decreased activity of CTP:choline-phosphate cytidylyltransferase (EC 2.7.7.15), or breakdown of phosphatidylcholine (13). The significance of increased PCho being associated with greater energy preservation after HI in our neonatal mice is not totally clear, but one can speculate that the origin is the repair of damage or resuming of normal growth. The relative strength of metabolite roles is also apparent when reading Figure 5b from left to right. The ATP–ADP resonance peak is by far the most important, maintaining dominance as models with more metabolites are allowed.
Although the NMR studies were performed in brain extracts, brains were removed after there was considerable time for significant metabolite exchanges between brain and blood. A recent search for biomarkers in blood taken ≤5 min after asphyxia in a nonhuman primate model of neonatal asphyxia (14) used 2D gas chromatography with time-of-flight mass spectrometry and found significant changes in several blood metabolites, including succinate, lactate, glutamate, malate, and leucine. Although these metabolites do not freely traverse an intact blood–brain barrier, further studies of blood metabolites in our mouse model would be needed to assess their contributions to the brain metabolites that we quantified.
What one would like to see, and which was not possible in this study, is metabolomics scores and loadings plots for later times after HI, ideally with one or more outcome scores associated with each point in the multidimensional metabolite space. In an ideal situation, a metabolomics approach would have different clusters for different insult severities early after HI, different clusters for different treatment groups, and best outcome scores associated very strongly with one or more treatment groups.
It is natural to ask when and how this study’s complex NMR methods and statistical analyses might become clinically relevant and if research paths using in vivo magnetic resonance spectroscopy (MRS) are appropriate at this time. A major limit at this time is the spectral resolution of in vivo MRS at available magnetic fields. In 2003, the US Food and Drug Administration specified that risks to neonates younger than 1 mo have not been ruled out above 4 Tesla, nor have they been ruled out above 8 Tesla for older children and adults (15). Nevertheless, impressive technological advances in 1H NMR metabolic profiling suggest a bright future. In a preclinical adult rodent study, 20 metabolites were accurately quantified in 14.1 Tesla in vivo 1H MRS (16). In an in vivo 4.0 Tesla human MRS clinical study of adults, the 2D J-resolved spectroscopy method was used to improve spectral resolution (17). Moreover, a totally new MRS approach is evolving based on dynamic nuclear polarization spectroscopy, a technology that is more complex and more expensive, but 35,000 times more sensitive than existing systems at NMR detection of 13C metabolites (18).
In summary, this study demonstrated an important role for metabolomics in distinguishing different treatment and outcome groups, and it has identified potentially important biomarkers. We are encouraged that roles will ultimately emerge for multivariate metabolomics in helping clinical management.
Methods
Animal Procedures: Induction of HI in Neonatal Mice and Mild Hypothermia
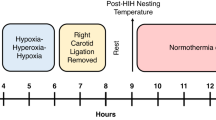
All animal experimental procedures were approved by the University of California, San Francisco institutional animal care and use committee. For each HI experiment, a litter of 10 CD1 mice of either sex (Charles River Laboratories, Wilmington, MA) underwent the Vannucci procedure of neonatal HI (19,20,21) on P7. Briefly, each pup was placed on a 37 °C temperature-controlled pad for general anesthesia from spontaneous respirations of 3.0% isoflurane in oxygen. After the right common carotid artery was permanently ligated, pups recovered from anesthesia on a warming pad with littermates before being returned to the dam for 90 min of feeding. HI was induced thereafter by placing the pups for 30 min into chambers that were maintained at 37 °C and through which a mixture of 8% oxygen and 92% nitrogen flowed. During this period, one pup’s temperature was monitored continuously by a thermistor temperature filament (outside diameter: 0.82 mm; probe type: RET-4; Physitemp Instruments, Clifton, NY) attached to the abdominal skin. Temperatures of all other pups were checked regularly using a noncontact infrared laser thermometer. After HI, pups were placed with the dam for 1 h of recovery. They were then exposed to 3.5 h of either hypothermia (31 °C) or normothermia (37 °C). During hypothermia or normothermia, pup core temperatures were monitored as described above. Pups in the normothermia group were returned to the dam for 24 h after HI. Those in the hypothermia group underwent a slow rewarming (0.1 °C per minute) to 37 °C, the temperature of the normothermia group, and were then returned to the dam.
At each predetermined time point listed below, two pups, one at a time, were decapitated under isoflurane anesthesia, after which the heads were placed in ice. The two brains, after being removed rapidly and then snap-frozen in liquid nitrogen, were saved for subsequent perchloric acid extraction and NMR analysis of the mixed extracts. The time interval between decapitation and snap-freezing was always <60 s, which resulted in significant preservation of ATP and other high-energy phosphates that are known to disappear quickly in the brains of mice after decapitation (22,23).
Using the notation P7 for postnatal day 7 pups and P8 for postnatal day 8 pups, animals were assigned to the following six groups, from which brains were taken for NMR analysis: (i) The HI group: P7 brains obtained immediately at the end of HI, without reoxygenation; (ii) the control group (control): no HI, no hypothermia; (iii) the P7 hypothermia group (P7 hypothermia): animals underwent the full HI protocol and hypothermia treatment, and brains were obtained immediately at the end of 3.5 h of hypothermia; (iv) The P7 normothermia group (P7 normothermia): animals underwent the full HI protocol and a 3.5 h normothermia treatment, at the end of which their brains were obtained; (v) the P8 hypothermia group (P8 hypothermia): animals followed the same procedure as those in the P7 hypothermia group but underwent a slow rewarming before returning to the dam, with brains being obtained 24 h after HI; and (vi) the P8 normothermia group (P8 normothermia): animals followed the same procedure as those in the P7 normothermia group but returned to the dam after treatment, with brains being obtained at 24 h after HI. For NMR analysis, a total of 36 pups were used. Three NMR tubes were obtained from six mice for each of the six groups.
Perchloric Acid Metabolite Extraction
Each perchloric acid extraction was obtained from two whole brains of sibling P7 mice in the same experimental group. Perchloric acid extraction was done as in earlier studies (24,25,26). In brief, to obtain extracts for one NMR tube for a given time point, two frozen brains of sibling P7 mice in the same experimental group and time point were ground together under liquid nitrogen in a precooled mortar and then extracted using 30 ml/g (dry mass) of ice-cold 12% perchloric acid. After centrifugation, each supernatant was neutralized with KOH to pH 7.0–7.2 and then centrifuged again. The final supernatant was filtered through a 0.22 μm sterile MCE Fisherbrand syringe filter (Fisher Scientific, Pittsburgh, PA) and lyophilized for 24 h on VirTis BenchTop 2K lyophilizer (SP Industries, Gardiner, NY).
NMR Data Acquisition and Analysis
Before NMR analysis, lyophilized dry extracts were weighed and dissolved in 280 μl of D2O containing 1 μmol/l 4,4-dimethyl-4-silapentane-1-sulfonic acid, a reference compound for NMR determinations of chemical shifts and signal intensities. Samples were neutralized to pD 7.0–7.2 with DCl and NaOD before being loaded into 3 mm NMR tubes (Select Series S-3-900-7; Norell, Landisville, NJ). Spectra were acquired at the Central California 900 MHz NMR Facility, which operates with National Institutes of Health support (GM68933) in the QB3 facilities at the University of California, Berkeley (Berkeley, CA). One-dimensional 1H spectra were acquired on a Bruker Avance II 900 MHz (21.1 Tesla; Bruker, Billerica, MA) NMR spectrometer equipped with a 5 mm CPTCI cryoprobe (Bruker) using a 30° tip angle pulse with 128 K points over 4.65 s and with a recycle delay of 1.0 s, for a total interexperiment delay of 5.65 s. No solvent suppression was used. Spectra were processed without apodization or zero filling and were referenced to 4,4-dimethyl-4-silapentane-1-sulfonic acid. The line widths of the methyl resonances of 4,4-dimethyl-4-silapentane-1-sulfonic acid typically ranged from 1.0 to 1.5 Hz (0.0011–0.0017 ppm).
A check on 1H resonance assignments for metabolites was done for each of the temperature conditions by performing 2D 1H J-resolved NMR spectroscopy (2D JRES spectroscopy), using acquisition methods described elsewhere (27). In a 2D JRES spectrum, units for the horizontal axis are ppm, and Hz units for the vertical axis are used to display distances of J-coupled peaks from each resonance’s center. Projection downward along the ppm axis consolidates the multiplets of each metabolite into a single peak, thereby greatly reducing the resonance peak overlap for different metabolites. The resulting projection of the 2D JRES plot is known as a pJRES 1H spectrum.
Figure 6a – c show the full spectral regions, comparing one-pulse data (top tracings) with one-dimensional pJRES data (bottom tracings). Figure 7 shows expanded views of two small spectral regions containing resonance peaks whose quantifications were important to the study. In Figure 7a , singlet resonance peaks from the adenosine moiety in ATP (8.49 ppm) and ADP (8.59 ppm) can be found. Quantification of the uncontaminated ADP was straightforward, but quantification of ATP involved deconvolution because of an overlapping component.
Using one nuclear magnetic resonance (NMR) tube with extracts from control mice, two different types of 1H NMR spectra were obtained sequentially without moving the sample or changing static magnetic fields. A standard one-pulse 1H NMR high-resolution 900 MHz spectrum (top tracing) is aligned above a 1H pJRES spectrum, the latter being a one-dimensional projection along the 1H ppm axis of a two-dimensional J-resolved 1H NMR plot, as discussed in the Methods. Notable differences in the pJRES spectra include its substantially flatter baseline and, for many metabolites, the consolidation of many resonance peak clusters into one peak. For the horizontal axis, parts-per-million (ppm) assignments relative to the DSS reference peak are from published chemical shifts. Spectral regions are (a) 0.8–2.9 ppm; (b) 2.9–4.2 ppm; (c) 5.6–8.8 ppm. Ala, alanine; Asp, aspartate; Cr, creatine; DSS, 4,4-dimethyl-4-silapentane-1-sulfonic acid; GABA, γ-aminobutyric acid; Gln, glutamine; Glu, glutamate; Gly, glycine; GPCho, glycerophosphocholine; His, histidine; Ile, isoleucine; Leu, leucine; NAA, N-acetyl-aspartate; NAAG, N-acetyl-aspartyl-glutamate; PCho, phosphocholine; PCr, phosphocreatine; Phe, phenylalanine; Thr, threonine; Trp, tryptophan; Tyr, tyrosine; Val, valine.
Expanded view of 1H resonance changes for high-energy phosphate metabolites in six experimental groups. (a) ATP and ADP are shown for a spectral region generally free of peaks from nucleotide phosphates containing guanosine, cytosine, and thymidine. (b) The spectral region showing PCr and Cr being clearly separated. Cr, creatine; PCr, phosphocreatine.
1H NMR metabolites corresponding to different chemical shifts were identified and preprocessed with TopSpin 3.1 and AMIX software (Bruker) and quantified with iNMR (Nucleomatica, Molfetta, Italy). All chemical shift assignments for peaks and clusters were checked against chemical shift assignments published in careful studies (28,29). Relaxation time corrections were not performed because the same NMR pulse sequence was used for all runs and the goal of the analysis was primarily to detect large differences in each metabolite from its control value. Metabolite signal intensities were computed relative to 4,4-dimethyl-4-silapentane-1-sulfonic acid and then normalized both to the weight of the dry powder that was mixed with D2O before NMR measurements and to the sum of quantifications of all metabolites shown in Figure 6 .
Brain Histology Assessment of Injury
Although it is known that mild hypothermia is therapeutic in HI mouse models of neonatal asphyxia (1), data were obtained to validate and quantify this for the exact conditions of our experiment. Twenty-four additional P7 mice underwent the same HI protocol already described, with 12 mice assigned randomly to each of the normothermia and hypothermia groups. Animals were euthanized 5 d after HI, and brains were perfused via intracardiac injection of buffered (0.1 mol/l phosphate, pH 7.2) 10% formalin freshly prepared from paraformaldehyde. Coronal sections were cut through the forebrain at 50-μm intervals with a Vibratome (Ted Pella, Redding, CA). Alternate sections were stained with cresyl violet for morphology, or with Perl’s stain to localize iron deposition. Brains were scored in a blinded manner as in earlier studies (20) for degree of injury, using a scale from 0 (no injury) to 24 (cystic infarct in multiple regions). Metabolomic data were not obtained.
Statistical and Metabolomic Analyses
Metabolite quantifications for all six experimental groups underwent within-group and between-group statistical analysis including pairwise t-testing, PCA, and the L1-penalized lasso regression analyses.
Pairwise comparison testing for differences in individual metabolites was performed via t-tests between groups treated with hypothermia and normothermia, as well as between those groups and control or HI groups. Therefore, there were 10 tests for each metabolite: P7 hypothermia vs. P7 normothermia; P8 hypothermia vs. P8 normothermia; control vs. P7 hypothermia; control vs. P7 normothermia; control vs. P8 hypothermia; control vs. P8 normothermia; HI vs. P7 hypothermia; HI vs. P7 normothermia; HI vs. P8 hypothermia; and HI vs. P8 normothermia. Multiplicity correction was effected using false-discovery rate methods (30).
In the PCA, done using SIMCA P+ v.13 software (Umetrics, San Jose, CA) as described previously (8,31), metabolite quantifications were centered and scaled to SD units and analyzed as vectors in a multivariate space, with one point (or one data vector) in the space corresponding to a full set of metabolite quantifications. Two-dimensional scores and loadings plots were made using the first and second principal components (PC1 and PC2), which correspond respectively to axes having the most and second-most variance for data vector projections. Scores plots were examined for separate groupings or clusterings of data from different treatment groups. Loadings plots, made from projecting individual metabolite vector components onto the plane of scores plots, were searched for the most important metabolites that contribute to clustering in the scores plots by examining metabolite components in the same quadrant as the clusters and maximally from the origin. Scores plots were assessed from two parameters, R2 and Q2, these being known respectively as the “explained variance” and the “cross-validation variance,” or that fraction of the total variance that can be attributed to the vector components in the scores plot (32).
Besides the scores and loadings plots, another PCA multivariate plot, the contribution plot, was used to compare metabolite differences between selected groups. This process starts with finding, metabolite by metabolite, each group’s average multivariate space vector for the chosen metabolite. For each metabolite, there is a vector subtraction of one group’s metabolite vector from the others, followed by a calculation of the resulting vector’s magnitude, which is then entered into the contribution plot.
A novel regression algorithm, the L1 penalized lasso (33,34), was employed as in our earlier studies to identify the most important metabolites in linear models that explain changes in PCr, which we used as a surrogate for ATP. The lasso (least absolute selection and shrinkage operator) has recently emerged as a forefront regression technique for situations in which the number of covariates (here metabolites) is large as compared with the number of observations. As explained previously, the lasso’s tuning parameter, λ, shrinks (reduces) the number of metabolites in the linear model as it increases upward from zero, initially eliminating noise variables, and ultimately eliminating the most important variables. The algorithm finds the optimal range for λ (35).
Median scores of brain injury for hypothermia and normothermia groups were calculated using Prism 5.0 (GraphPad Software, La Jolla, CA). The Mann–Whitney U-test was used to determine significance.
Statement of Financial Support
This research was supported by grants from the National Institutes of Health: R01-GM36747 and P41-GM68933 from the National Institute of General Medical Sciences and R01-NS33997 from the National Institute of Neurological Disorders and Stroke.
Disclosure
The authors have no financial ties to products in the study or potential/perceived conflicts of interest.
References
Carlsson Y, Wang X, Schwendimann L, et al. Combined effect of hypothermia and caspase-2 gene deficiency on neonatal hypoxic-ischemic brain injury. Pediatr Res 2012;71:566–72.
Sutcliffe IT, Smith HA, Stanimirovic D, Hutchison JS . Effects of moderate hypothermia on IL-1 beta-induced leukocyte rolling and adhesion in pial microcirculation of mice and on proinflammatory gene expression in human cerebral endothelial cells. J Cereb Blood Flow Metab 2001;21:1310–9.
Tsuchiya D, Hong S, Suh SW, Kayama T, Panter SS, Weinstein PR . Mild hypothermia reduces zinc translocation, neuronal cell death, and mortality after transient global ischemia in mice. J Cereb Blood Flow Metab 2002;22:1231–8.
Xiang Z, Thomas S, Pasinetti G . Increased neuronal injury in transgenic mice with neuronal overexpression of human cyclooxygenase-2 is reversed by hypothermia and rofecoxib treatment. Curr Neurovasc Res 2007;4:274–9.
Griffin JL, Cemal CK, Pook MA . Defining a metabolic phenotype in the brain of a transgenic mouse model of spinocerebellar ataxia 3. Physiol Genomics 2004;16:334–40.
Pears MR, Cooper JD, Mitchison HM, Mortishire-Smith RJ, Pearce DA, Griffin JL . High resolution 1H NMR-based metabolomics indicates a neurotransmitter cycling deficit in cerebral tissue from a mouse model of Batten disease. J Biol Chem 2005;280:42508–14.
Salek RM, Colebrooke RE, Macintosh R, et al. A metabolomic study of brain tissues from aged mice with low expression of the vesicular monoamine transporter 2 (VMAT2) gene. Neurochem Res 2008;33:292–300.
Liu JL, Wang HL, Zhang LF, et al. Metabonomics study of brain-specific human S100B transgenic mice by using high-performance liquid chromatography coupled with quadrupole time of flight mass spectrometry. Biol Pharm Bull 2011;34:871–6.
Bak LK, Johansen ML, Schousboe A, Waagepetersen HS . Among the branched-chain amino acids, only valine metabolism is up-regulated in astrocytes during glutamate exposure. J Neurosci Res 2007;85:3465–70.
Cole JT, Sweatt AJ, Hutson SM . Expression of mitochondrial branched-chain aminotransferase and a-keto-acid dehydrogenase in rat brain: implications for neurotransmitter metabolism. Front Neuroanat 2012;6:18.
Ten VS, Starkov A . Hypoxic-ischemic injury in the developing brain: the role of reactive oxygen species originating in mitochondria. Neurol Res Int 2012;2012:542976.
Barks JD, Liu YQ, Shangguan Y, Silverstein FS . Phenobarbital augments hypothermic neuroprotection. Pediatr Res 2010;67:532–7.
Geddes JW, Panchalingam K, Keller JN, Pettegrew JW . Elevated phosphocholine and phosphatidylcholine following rat entorhinal cortex lesions. Neurobiol Aging 1997;18:305–8.
Beckstrom AC, Humston EM, Snyder LR, Synovec RE, Juul SE . Application of comprehensive two-dimensional gas chromatography with time-of-flight mass spectrometry method to identify potential biomarkers of perinatal asphyxia in a non-human primate model. J Chromatogr A 2011;1218:1899–1906.
Guidance for Industry and FDA Staff: Criteria for Significant Risk Investigations of Magnetic Resonance Diagnostic Devices. In: U.S. Department of Health and Human Services FaDA, Center for Devices and Radiological Health, 2003:1–5.
Mlynárik V, Cudalbu C, Xin L, Gruetter R . 1H NMR spectroscopy of rat brain in vivo at 14.1Tesla: improvements in quantification of the neurochemical profile. J Magn Reson 2008;194:163–8.
Silveri MM, Jensen JE, Rosso IM, Sneider JT, Yurgelun-Todd DA . Preliminary evidence for white matter metabolite differences in marijuana-dependent young men using 2D J-resolved magnetic resonance spectroscopic imaging at 4 Tesla. Psychiatry Res 2011;191:201–11.
Gallagher FA, Kettunen MI, Day SE, et al. Detection of tumor glutamate metabolism in vivo using (13)C magnetic resonance spectroscopy and hyperpolarized [1-(13)C]glutamate. Magn Reson Med 2011;66:18–23.
Vannucci RC . Cerebral carbohydrate and energy metabolism in perinatal hypoxic-ischemic brain damage. Brain Pathol 1992;2:229–34.
Sheldon RA, Sedik C, Ferriero DM . Strain-related brain injury in neonatal mice subjected to hypoxia-ischemia. Brain Res 1998;810:114–22.
Sheldon RA, Osredkar D, Lee CL, Jiang X, Mu D, Ferriero DM . HIF-1 alpha-deficient mice have increased brain injury after neonatal hypoxia-ischemia. Dev Neurosci 2009;31:452–8.
Seyfried FJ, Adachi N, Arai T . Suppression of energy requirement by lidocaine in the ischemic mouse brain. J Neurosurg Anesthesiol 2005;17:75–81.
Thurston JH, McDougal DB Jr . Effect of ischemia on metabolism of the brain of the newborn mouse. Am J Physiol 1969;216:348–52.
Espanol MT, Litt L, Yang GY, et al. Tolerance of low intracellular pH during hypercapnia by rat cortical brain slices: a 31P/1H NMR study. J Neurochem 1992;59:1820–8.
Liu J, Hirai K, Litt L . Fructose-1,6-bisphosphate does not preserve ATP in hypoxic-ischemic neonatal cerebrocortical slices. Brain Res 2008;1238:230–8.
Liu J, Segal M, Yoo S, et al. Antioxidant effect of ethyl pyruvate in respiring neonatal cerebrocortical slices after H(2)O(2) stress. Neurochem Int 2009;54:106–10.
Ludwig C, Viant MR . Two-dimensional J-resolved NMR spectroscopy: review of a key methodology in the metabolomics toolbox. Phytochem Anal 2010;21:22–32.
Govindaraju V, Young K, Maudsley AA . Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed 2000;13:129–53.
Willker W, Engelmann J, Brand A, Leibfritz D . Metabolite identification in cell extracts and culture media by proton-detected 2D [1H, 13C] NMR spectroscopy. J Magn Res Anal 1996;2:21–32.
Storey JD . A direct approach to false discovery rates. J Roy Stat Soc Ser B Stat Methodol 2002;64:479–98.
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S . Multi- and Megavariate Data Analysis Part I: Basic Principles and Applications. Second Revised and Enlarged Edition. Umeå, Sweden: MKS Umetrics AB 2006:1–103.
Eriksson L, Antti H, Gottfries J, et al. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm). Anal Bioanal Chem 2004;380:419–29.
Efron B, Hastie T, Johnstone I, Tibshirani R . Least angle regression. Ann Statist 2004;32:407–99.
Tibshirani R . Regression shrinkage and selection via the lasso. J Roy Stat Soc Ser B Stat Methodol 1996;58:267–88.
Hastie T, Tibshirani R, Friedman J . The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd edn. New York: Springer, 2010:61–94.
Acknowledgements
Gratefully acknowledged are very helpful and inspiring discussions with Ulrich Günther, Mark Viant, and Christian Ludwig, along with their hospitality during visits to the University of Birmingham’s Henry Wellcome Center for Biomolecular Spectroscopy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Liu, J., Sheldon, R., Segal, M. et al. 1H nuclear magnetic resonance brain metabolomics in neonatal mice after hypoxia–ischemia distinguished normothermic recovery from mild hypothermia recoveries. Pediatr Res 74, 170–179 (2013). https://doi.org/10.1038/pr.2013.88
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2013.88