Abstract
Background:
Celiac disease (CD) has been associated with HLA class II heterodimers. This study aimed at determining the HLA genotypic and allelic distribution in Greek children with CD as compared with the general population.
Methods:
A total of 118 children with CD and 120 healthy individuals serving as controls were included in the study.
Results:
Higher frequencies for HLA-DQB1*02:01 (40.25 vs. 9.58%, P < 0.001) and DQB1*02:02 (20.34 vs. 5.42%, P < 0.001) were observed in patients with CD, whereas HLA-DQB1*03:01 (16.53 vs. 30.42%, P < 0.001), DQB1*05:01 (0.85 vs. 10%, P < 0.001), and DQB1*05:02 (5.51 vs. 17.92%, P < 0.001) were significantly lower, as compared with the controls. DQA1*02:01 (patients with CD vs. controls: 20.76 vs. 6.67%, P < 0.001) and DQA1*05:01 (40.25 vs. 9.58%, P < 0.001) were significantly more frequent in patients. The frequencies of HLA-DQA1* 01:01, *01:02, *01:04, and *05:05 were significantly lower in patients (P < 0.001). The haplotype mainly associated with CD was DRB1*03-DQB1*02:01-DQA1*05:01; patients with CD vs. controls: 39.83 vs. 9.58%, P < 0.001. In total, 84.75% of patients carried DQ2 (vs. 21.67% in controls, P < 0.001), whereas 11.02% were DQ8 positive/DQ2 negative.
Conclusion:
This study confirms the existing data and provides additional evidence supporting a strong genetic predisposition for CD associated with the class II alleles DQB1*02 and DQA1*05 encoding the serological specificity DQ2.
Similar content being viewed by others
Main
Celiac disease (CD) is a chronic intestinal inflammatory disorder resulting in villous atrophy and flattening of the mucosa. It is activated in genetically susceptible individuals by the dietary ingestion of proline- and glutamine-rich proteins that are found in wheat, rye, and barley and are widely termed “gluten” (1). CD can present at any age with a variety of symptoms ranging from the classical CD, characterized by chronic diarrhea, abdominal distention, and failure to thrive within the first couple of years of life, to the atypical, with extraintestinal manifestations such as unexplained iron deficiency anemia, short stature, osteoporosis, and pubertal delay (2,3,4). Recent epidemiologic studies have revealed that positive serological tests for CD can be found up to 1% of the general population (5).
Although the exact genetic basis of the disease has not been determined, certain individuals appear to be genetically predisposed to develop CD. The markers that have particularly been associated with CD are the HLA-DQ2 and/or DQ8 heterodimers. This association can be explained by the fact that gluten peptides can be presented in HLA-DQ2 and DQ8 molecules on antigen-presenting cells. Gluten-specific CD4+ T cells in the lamina propria respond to these peptides, and this probably enhances cytotoxicity of intraepithelial lymphocytes against the intestinal epithelium (6).
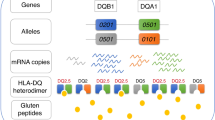
HLA-DQ (DQ) is a cell surface receptor–type protein found on antigen-presenting cells. It is encoded on the HLA region of chromosome 6p21.3, which is historically known as the “D” antigen region. This region encodes the subunits DP, DQ, and DR, which are the major histocompatibility complex class II antigens in humans. DQ is made up of two different subunits to form a αβ-heterodimer. Each subunit is encoded by its own gene. The α subunit is encoded by the HLA-DQA1 gene and the β subunit is encoded by the HLA-DQB1 gene. Both loci are characterized by extensive genetic variability in the human population. The DQ loci are in close genetic linkage with HLA-DR but are less closely linked to HLA-DP, HLA-A, HLA-B, and HLA-C.
The HLA-DQ2 heterodimer is formed by one β2 chain encoded by the HLA-DQB1*02 allele (DQB1*02:01 or 02:02) and one α5-chain encoded by the HLA-DQA1*05 allele (DQA1*05:01 or 05:05). Most patients with CD carry DQB1*02:01-DQA1*05:01 on a DRB1*03 haplotype (DRB1*03-DQB1*02:01-DQA1*05:01). However, the α5 and β2 chains of the DQ2 heterodimer may also be encoded in trans with the DQA1*05:05 allele, usually on DRB1*11 or 12 or 13 haplotypes, and the DQB1*02:02 allele, usually on a DRB1*07 haplotype (i.e., DRB1*11 or 12 or 13-DQB1*03:01-DQA1*05:05/DRB1*07-DQB1*02:02-DQA1*02:01 genotype) (7,8,9,10,11). This HLA-DQ2 heterodimer is present in at least 90% of patients with CD, although a very small number of patients with CD have been reported carrying only one chain of the DQ2 heterodimer (12).
The few patients with CD who are DQ2-negative, as well as a very small portion of those who are HLA-DQ2 positive, frequently have the DQ8 heterodimer. The HLA-DQ8 heterodimer is formed by one β chain and one α chain encoded by the HLA-DQB1*03:02 and HLA-DQA1*03 alleles, respectively. These two alleles, along with the DRB1*04 allele, form the DR4-DQ8 haplotype (DRB1*04-DQB1*03:02-DQA1*03:01) (13,14). More than 95% of patients with CD carry HLA-DQ2 and/or HLA-DQ8, and hence the absence of these molecules reduces significantly the likelihood of CD (15).
In this study, we aimed at assessing the HLA-DR/DQ genotypic and allelic distribution in a large cohort of children with CD in Greece, as compared with the general population, as well as evaluating the HLA effect on disease susceptibility.
Results
Allelic Distribution
The overall distribution of HLA DQB1 and HLA DQA1 alleles significantly differed between patients and controls (Pearson’s χ2, both P values <0.001). The frequencies of all HLA DQB1 and HLA DQA1 alleles in our CD group and comparisons with the general population are shown in Tables 1 and 2 . With regard to HLA DQB1 alleles, significantly higher prevalence in patients with CD was found for DQB1*02:01 (P < 0.001) and DQB1*02:02 (P < 0.001). In contrast, HLA-DQB1*03:01 (P < 0.001), DQB1*05:01 (P < 0.001), and DQB1*05:02 (P < 0.001) were significantly lower in patients with CD. Among HLA-DQA1 alleles, DQA1*02:01 (P < 0.001) and DQA1*05:01 (P < 0.001) were significantly more frequent in patients, whereas the observed frequencies of HLA-DQA1* 01:01, 01:02, 01:04, and 05:05 in patients were significantly lower than those in controls (P < 0.001). Table 3 shows the HLA-DRB1 allelic distribution. Significantly higher frequencies of DRB1*03 (P < 0.001) and DRB1*07 (P < 0.001) were found in patients with CD as compared with controls, whereas HLA-DRB1*01 and DRB1*16 presented much lower frequencies in the patients group (P < 0.001 and P < 0.001, respectively).
Haplotypic Distribution
In total, the HLA DRB1-DQB1-DQA1 haplotypic distribution was significantly different in patients with CD as compared with controls (Pearson’s χ2, P value <0.001). Individual comparisons are listed in Table 4 . The DRB1*03-DQB1*02:01-DQA1*05:01 and DRB1*07-DQB1*02:02-DQA1*02:01 haplotypes were significantly more common in patients as compared with controls (both P values <0.001). However, the observed frequency of DRB1*11/12/13-DQB1*03:01-DQA1*05:05 in patients was lower than that in control subjects (P < 0.001). The frequency of the DRB1*04-DQB1*03:02-DQA1*03:01 haplotype in our CD group was not significantly different from controls.
Genotypic Distribution
The genotypic distribution, as defined by the combination of haplotypes, appeared to be significantly divergent between patients and controls (Pearson’s χ2, P value <0.001). Detailed comparisons are illustrated in Table 5 .
In the whole data set, 95.8% of individuals with CD were DQ2 and/or DQ8 positive (4.2% DQ2 and DQ8 negative), as compared with 32.5% in the general population (P < 0.001). In greater detail, 84.8% of patients were DQ2 positive (vs. 21.7% in controls, P < 0.001). Among those, the following subclasses were observed: DRB1*03-DQB1*02:01-DQA1*05:01 in homozygosis 13.6% vs. 1.7% (P = 0.001), with DRB1*07-DQB1*02:02-DQA1*02:01 17.8% vs. 1.67% (P < 0.001), with DRB1*04-DQB1*03:02-DQA1*03:01 3.4% vs. 0% (not significant (NS)), in trans DRB1*11 or 12 or 13-DQB1*03:01-DQA1*05:05/DRB1*07-DQB1*02:02-DQA1*02:01 17.8% vs. 4.2% (P = 0.001), with DRB1*11/12/13-DQB1*03:01-DQA1*05:05 9.3% vs. 5.8% (NS), and DRB1*03-DQB1*02:01-DQA1*05:01 with other haplotypes 22.9% vs. 8.3% (P = 0.002).
Eleven percent of patients were DQ8 positive/DQ2 negative (vs. 10.83% in controls, NS). In homozygosis, DRB1*04-DQB1*03:02-DQA1*03:01/DRB1*04-DQB1*03:02-DQA1*03:01 1.7% vs. 0% (NS) and with other haplotypes DRB1*04-DQB1*03:02-DQA1*03:01 /x 9.3% vs. 10.8% (NS). After stratification according to DQ2 positivity, DQ8 was found significantly more frequently in CD-DQ2-negative patients as compared with healthy DQ2-negative controls (72.2% vs. 13.8%, respectively, P < 0.001). In DQ2-positive individuals, there was no difference in DQ8 positivity (P = 0.6).
Among the five DQ2/DQ8-negative individuals with CD (4.3% in patients vs. 67.5% in controls, P < 0.001), four carry half of the DQ2 molecule. Three patients carried DQB1*02 in the absence of DQA1*05, which results in carrying the β2 chain without the α5 chain (one patient was DRB1*07-DQB1*02:02-DQA1*02:01 homozygous and two patients were DRB1*07-DQB1*02:02-DQA1*02:01 heterozygous), and one subject had DQA1*05 in the absence of DQB1*02 (DRB1*11-DQB1*03:01-DQA1*05:05 homozygous), which also results in carrying half the DQ2 molecule (α5 chain without the β2 chain). Finally, the fifth DQ2/DQ8-negative patient was negative for both DQ2 chains (α5 and β2) and carried the DRB1*14-DQB1*05:03-DQA1*01:04/DRB1*16-DQB1*05:02-DQA1*01:02 genotype. This corresponds to a 1/118 (0.8%) probability of combined DQ2/DQ8/α5/β2 negativity in CD patients.
Discussion
CD is a permanent gluten intolerance strongly associated with HLA class II antigens. The overpresentation of particular HLA alleles and haplotypes has been described in several populations (15,16). The strong genetic predisposition was quantified by Bourgey et al. (17), who estimated that the overall risk that a sib of a patient with CD will develop the disease is ~10%, ranging from 0.1% to 29% when HLA-DQ information of the proband, parents, and sib was considered. Megiorni et al. (18) obtained a risk gradient ranging from 1:7 for DQ2 and DQ8 individuals down to 1:2,518 for subjects lacking all predisposing HLA-DQ factors. In this study, we provide data, for the first time in a large Greek pediatric population from a single center, supporting a strong association between HLA-DQB1, HLA-DQA1, and HLA-DRB1 and CD. Overall, we found that 95.8% of individuals with CD were DQ2 and/or DQ8 positive, as compared with 32.5% of the healthy individuals, a result similar to those in other Caucasian populations (15,16,19). This result is very important in terms of diagnostic policies in Greece, given that DQ2 and/or DQ8 positivity holds a significant role in the latest European Society of Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of CD (20). The majority of DQ2-positive patients were found to be homozygous or heterozygous for the DRB1*03-DQB1*02:01-DQA1*05:01 haplotype (DR3-DQ2). However, the likelihood of DQ2 positivity (84.8%) is clearly lower than the 90–95% reported in a Northern European population (21). This could be attributed to the remarkably lower frequency of the DQA1*05:05-carrying haplotypes (DRB1*11/12/13-DQB1*03:01-DQA1*05:05) in patients as compared with controls. In populations in which the DQ2 positivity is >90%, the distribution of the DQA1*05:05-carrying haplotypes (DRB1*11/12/13-DQB1*03:01-DQA1*05:05) is similar between patients and controls. This could explain the slightly lower frequency of DQ2-positive individuals with CD in our population, because among the haplotypes that are necessary for a DQ2 heterodimer, only the aforementioned haplotypes were found more frequently in controls in our study. The proportion of patients carrying the DRB1*11/12/13-DQB1*03:01-DQA1*05:05 haplotypes (formerly known as DR5-DQ7) has been shown to vary across Europe, not only in patients but also in the healthy population as well, with frequencies reported from Spain (19) and Italy (11) being higher as compared with northern Europe.
Among the five DQ2- and DQ8-negative patients, three of them carried the haplotype DRB1*07-DQB1*02:02-DQA1*02:01 (DR7-DQ2) without DQA1*05 (i.e., had the β2 chain without the α5 chain) and one patient carried the DRB1*11-DQB1*03:01-DQA1*05:05 (DR5-DQ7) haplotype in homozygosis (i.e., had the α5 chain without the β2 chain). The fifth patient lacked all HLA-susceptibility factors and was a girl with short stature, positive serological test, and partial villous atrophy who responded to a gluten-free diet. She carried the DRB1*14-DQB1*05:03-DQA1*01:04/DRB1*16-DQB1*05:02-DQA1*01:02 genotype. In a multicenter study conducted in Europe in 2002, 61 of the 1,008 patients with CD lacked the DQ2 or DQ8 heterodimer (6.0%), 57 of them (5.6%) encoded one half of the DQ2 heterodimer, and 4 (0.4%) were DQ2, DQ8, α5, β2 negative (15). Our results are similar to those reported from the other studies. These findings demonstrate that just one of the alleles DQA1*05 or DQB1*02, coding for half of the DQ2 heterodimer molecule, would confer a predisposition toward triggering CD (15,22).
In our CD population, we have found a pronounced increase of the DQB1*02:01, DQB1*02:02, DQA1*02:01, DQA1*05:01, DRB1*03, and DRB1*07 alleles, which are those classically associated with CD in other populations as well (23,24,25,26,27). No association was detected with DRB1*04, DQB1*03:02, and DQA1*03:01 alleles, an observation comparable with results reported by others (19). In our series, the observed frequencies of DQA1*01:01, DQA1*01:02, DQA1*01:04, DQA1*05:05, DQB1*03:01, DQB1*05:01, DQB1*05:02, DRB1*01, and DRB1*16 in patients with CD are lower than those in controls. It remains unclear whether this negative association confers a real protective effect against the development of CD.
The DQ2 and/or DQ8 positivity ratio in our study was 95.8%/32.5% (patients with CD vs. controls), a finding comparable to those reported in other Caucasians. It is interesting that despite the lower proportion of DQ2-positive patients, the relatively higher percentage of DQ8 positivity, as compared with other CD populations, allows the final combined DQ2/DQ8 positivity to marginally exceed 95%, a level frequently observed in other studies.
A number of functional studies have recently focused on the role of a single β2 chain encoded by DRB1*07-DQB1*02:02-DQA1*02:01 in the absence of DRB1*03-DQB1*02:01-DQA1*05:01 in the pathogenesis of CD. Hernandez-Charro et al. suggested that the DRB1*07-DQA1*02:01-DQB1*02:02 haplotype does not confer a significantly higher risk of CD, unless it is expressed with DRB1*03-DQA1*05-DQB1*02. This hypothesis is confirmed in our study by the significantly higher relative risk for CD in the presence of DRB1*07-DQA1*02:01-DQB1*02:02/DRB1*03-DQA1*05-DQB1*02 (odds ratio: 12.8, 95% confidence interval: 2.9–55.9) as compared with the relative risk derived from the combination of DRB1*07-DQA1*02:01-DQB1*02:02 with any other haplotype (odds ratio: 3.1, 95% confidence interval: 1.4–6.8).
It is noteworthy that, although, most patients with CD carry the HLA DQ2 or HLA DQ8 heterodimer, the HLA susceptibility genes account for only about 40% of the whole genetic risk. The challenge now is to explain the remaining 60% (28,29), which could be attributed to additional genetic factors such as CTLA-4, a non-HLA gene thought to regulate T-cell immune function (30).
As recommended, HLA types should not be classified only as DQ2/DQ8 positive or negative. Clinicians involved in the diagnosis of CD must also consider the presence of one half of the DQ2 heterodimer determined by genetic analysis (15).
In conclusion, this study confirms and extends evidence for a strong genetic predisposition for CD associated with the class II alleles DQB1*02-DQA1*05 encoding the DQ2 molecule in the pediatric Greek population. The main associated haplotype was DRB1*03-DQB1*02:01-DQA1*05:01, followed by DRB1*07-DQB1*02:02-DQA1*02:01. The frequency of the haplotype DRB1*04-DQB1*03:02-DQA1*03:01 was not significantly higher in patients vs. controls. Among DQ2-negative individuals, DQ8 was found significantly more frequently in patients with CD than in controls.
Methods
Subjects
This study was carried out in the First Department of Pediatrics in the University of Athens, Greece, following approval by the review board of “Aghia Sophia” Children’s Hospital. Written, informed consent was obtained from the legal guardians of all participating children. Any child of Greek origin diagnosed with CD according to the criteria proposed by the European Society of Pediatric Gastroenterology, Hepatology, and Nutrition was eligible for inclusion (31). For children diagnosed prior to 1990, the “Interlaken criteria” were applied, whereas for those diagnosed after 1990, the revised European Society of Pediatric Gastroenterology, Hepatology, and Nutrition criteria were adopted. Finally, a total of 118 patients were included; a panel of 120 healthy individuals, blood and bone marrow donors of the same origin, served as controls.
HLA Typing
Samples of peripheral blood were taken from patients and controls and genomic DNA was extracted using an automated system (MagNA Pure Compact, Roche, Basel, Switzerland). HLA class II (DRB1, DQA1, DQB1 loci) genotyping was performed by sequence-specific primer PCR (Life Technologies, Carlsbad, CA; One Lambda, Canoga Park, CA; Olerup International AB, Stockolm, Sweden) and sequence-specific oligonucleotide primed PCR (Life Technologies; One Lambda). All the technical parts of the study were carried out in the Department of Immunology and Histocompatibility of “Aghia Sophia” Children’s Hospital. The haplotypes were deduced by means of the known patterns of linkage disequilibrium in the Greek population (32,33).
Statistical Analysis
Allelic, genotypic, and haplotypic frequencies in patients with CD and controls are presented as absolute (n) and relative frequencies (%). For the comparison of the allelic distribution for each genetic locus (DQA1, DQB1, and DRB1) between patients with CD and controls, 2 × 12, 2 × 16, and 2 × 13 tables were constructed, respectively, and Pearson’s χ2 test was applied using 0.05 as level of significance. To identify which allele differed significantly, in each case, between patients and controls, 12, 16, and 13 2 × 2 tables were formed (for DQA1, DQB1, and DRB1, respectively). Each table compared the distribution of a particular allele against all the others in patients and controls, using Fisher’s exact test. Due to the large number of comparisons, the level of statistical significance was adjusted accordingly (0.05/12 = 0.004 for DQA1, 0.05/16 = 0.003 for DQB1, and 0.05/13 = 0.0038 for DRB1). All relative risks were quantified by logistic regression analysis and expressed as odds ratios.A similar approach was adopted for the comparison of haplotypes and genotypes, with the exception of all haplotypes not coding for either the α or β chain of DQ2 and DQ8, which were grouped together. The same classification was applied for the genotypes resulting from these haplotypes. All statistical analyses were performed with Stata 11.0 MP statistical software (Stata Corp, College Station, TX)
Statement of Financial Support
No financial assistance was received to support this study.
References
Kagnoff MF . Overview and pathogenesis of celiac disease. Gastroenterology 2005;128:4 Suppl 1:S10–8.
Treem WR . Emerging concepts in celiac disease. Curr Opin Pediatr 2004;16:552–9.
Hill ID, Dirks MH, Liptak GS, et al. Guideline for the diagnosis and treatment of celiac disease in children: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr 2005;40:1–19.
Madani S, Kamat D . Clinical guidelines for celiac disease in children: what does it mean to the pediatrician/family practitioner? Clin Pediatr (Phila) 2006;45:213–9.
Catassi C, Kryszak D, Louis-Jacques O, et al. Detection of Celiac disease in primary care: a multicenter case-finding study in North America. Am J Gastroenterol 2007;102:1454–60.
Tjon JM, van Bergen J, Koning F . Celiac disease: how complicated can it get? Immunogenetics 2010;62:641–51.
Ek J, Albrechtsen D, Solheim BG, Thorsby E . Strong association between the HLA-Dw3-related B cell alloantigen -DRw3 and coeliac disease. Scand J Gastroenterol 1978;13:229–33.
Keuning JJ, Peña AS, van Leeuwen A, van Hooff JP, va Rood JJ . HLA-DW3 associated with coeliac disease. Lancet 1976;1:506–8.
Mearin ML, Biemond I, Peña AS, et al. HLA-DR phenotypes in Spanish coeliac children: their contribution to the understanding of the genetics of the disease. Gut 1983;24:532–7.
Tosi R, Vismara D, Tanigaki N, et al. Evidence that celiac disease is primarily associated with a DC locus allelic specificity. Clin Immunol Immunopathol 1983;28:395–404.
Trabace S, Giunta A, Rosso M, et al. HLA-ABC and DR antigens in celiac disease. A study in a pediatric Italian population. Vox Sang 1984;46:102–6.
van Heel DA, Hunt K, Greco L, Wijmenga C . Genetics in coeliac disease. Best Pract Res Clin Gastroenterol 2005;19:323–39.
Louka AS, Sollid LM . HLA in coeliac disease: unravelling the complex genetics of a complex disorder. Tissue Antigens 2003;61:105–17.
Mazzarella G, Maglio M, Paparo F, et al. An immunodominant DQ8 restricted gliadin peptide activates small intestinal immune response in in vitro cultured mucosa from HLA-DQ8 positive but not HLA-DQ8 negative coeliac patients. Gut 2003;52:57–62.
Karell K, Louka AS, Moodie SJ, et al. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol 2003;64:469–77.
Hernández-Charro B, Donat E, Miner I, Aranburu E, Sánchez-Valverde F, Ramos-Arroyo MA . Modifying effect of HLA haplotypes located trans to DQB1*02-DRB1*03 in celiac patients of Southern Europe. Tissue Antigens 2008;71:213–8.
Bourgey M, Calcagno G, Tinto N, et al. HLA related genetic risk for coeliac disease. Gut 2007;56:1054–9.
Megiorni F, Mora B, Bonamico M, et al. HLA-DQ and risk gradient for celiac disease. Hum Immunol 2009;70:55–9.
Donat E, Planelles D, Capilla-Villanueva A, Montoro JA, Palau F, Ribes-Koninckx C . Allelic distribution and the effect of haplotype combination for HLA type II loci in the celiac disease population of the Valencian community (Spain). Tissue Antigens 2009;73:255–61.
Husby S, Koletzko S, Korponay-Szabó IR, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr 2012;54:136–60.
Margaritte-Jeannin P, Babron MC, Bourgey M, et al. HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease. Tissue Antigens 2004;63:562–7.
Kagnoff MF . Celiac disease: pathogenesis of a model immunogenetic disease. J Clin Invest 2007;117:41–9.
Congia M, Frau F, Lampis R, et al. A high frequency of the A30, B18, DR3, DRw52, DQw2 extended haplotype in Sardinian celiac disease patients: further evidence that disease susceptibility is conferred by DQ A1*0501, B1*0201. Tissue Antigens 1992;39:78–83.
Herrera M, Theiler G, Augustovski F, et al. Molecular characterization of HLA class II genes in celiac disease patients of Latin American Caucasian origin. Tissue Antigens 1994;43:83–7.
Michalski JP, McCombs CC, Arai T, et al. HLA-DR, DQ genotypes of celiac disease patients and healthy subjects from the West of Ireland. Tissue Antigens 1996;47:127–33.
Tighe MR, Hall MA, Ashkenazi A, Siegler E, Lanchbury JS, Ciclitira PJ . Celiac disease among Ashkenazi Jews from Israel. A study of the HLA class II alleles and their associations with disease susceptibility. Hum Immunol 1993;38:270–6.
Tighe MR, Hall MA, Barbado M, Cardi E, Welsh KI, Ciclitira PJ . HLA class II alleles associated with celiac disease susceptibility in a southern European population. Tissue Antigens 1992;40:90–7.
Green PH, Cellier C . Celiac disease. N Engl J Med 2007;357:1731–43.
Rostom A, Murray JA, Kagnoff MF . American Gastroenterological Association (AGA) Institute technical review on the diagnosis and management of celiac disease. Gastroenterology 2006;131:1981–2002.
Djilali-Saiah I, Schmitz J, Harfouch-Hammoud E, Mougenot JF, Bach JF, Caillat-Zucman S . CTLA-4 gene polymorphism is associated with predisposition to coeliac disease. Gut 1998;43:187–9.
Revised criteria for diagnosis of coeliac disease. Report of Working Group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child 1990;65:909–11.
Pedron B, Yakouben K, Adjaoud D, et al. Listing of common HLA alleles and haplotypes based on the study of 356 families residing in the Paris, France, area: implications for unrelated hematopoietic stem cell donor selection. Hum Immunol 2005;66:721–31.
Papassavas EC, Spyropoulou-Vlachou M, Papassavas AC, Schipper RF, Doxiadis IN, Stavropoulos-Giokas C . MHC class I and class II phenotype, gene, and haplotype frequencies in Greeks using molecular typing data. Hum Immunol 2000;61:615–23.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Krini, M., Chouliaras, G., Kanariou, M. et al. HLA class II high-resolution genotyping in Greek children with celiac disease and impact on disease susceptibility. Pediatr Res 72, 625–630 (2012). https://doi.org/10.1038/pr.2012.133
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2012.133
This article is cited by
-
Distribution of celiac disease predisposing genes HLA-DQ2 and HLA-DQ8 in the native population of southern India
Indian Journal of Gastroenterology (2022)
-
Rosacea, Germs, and Bowels: A Review on Gastrointestinal Comorbidities and Gut–Skin Axis of Rosacea
Advances in Therapy (2021)
-
HLA-DQ genetics in children with celiac disease: a meta-analysis suggesting a two-step genetic screening procedure starting with HLA-DQ β chains
Pediatric Research (2018)
-
Novel genetic risk variants for pediatric celiac disease
Human Genomics (2016)
-
Assessment of the Genetic Basis of Rosacea by Genome-Wide Association Study
Journal of Investigative Dermatology (2015)