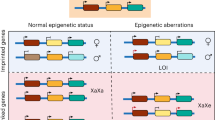
Abstract
To study the role of parental imprinting in human embryogenesis, we generated parthenogenetic human induced pluripotent stem cells (iPSCs) with a homozygote diploid karyotype. Global gene expression and DNA methylation analyses of the parthenogenetic cells enabled the identification of the entire repertoire of paternally expressed genes. We thus demonstrated that the gene U5D, encoding a variant of the U5 small RNA component of the spliceosome, is an imprinted gene. Introduction of the U5D gene into parthenogenetic cells partially corrected their molecular phenotype. Our analysis also uncovered multiple miRNAs existing as imprinted clustered transcripts, whose putative targets we then studied further. Examination of the consequences of parthenogenesis on human development identified marked effects on the differentiation of extraembryonic trophectoderm and embryonic liver and muscle tissues. This analysis suggests that distinct regulatory imprinted small RNAs and their targets have substantial roles in human development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Park, I.H. et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008).
McGrath, J. & Solter, D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 (1984).
Surani, M.A. & Barton, S.C. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science 222, 1034–1036 (1983).
Revazova, E.S. et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells 9, 432–449 (2007).
Mai, Q. et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res. 17, 1008–1019 (2007).
Brevini, T.A. et al. Cell lines derived from human parthenogenetic embryos can display aberrant centriole distribution and altered expression levels of mitotic spindle check-point transcripts. Stem Cell Rev. 5, 340–352 (2009).
Kim, K. et al. Histocompatible embryonic stem cells by parthenogenesis. Science 315, 482–486 (2007).
Kim, K. et al. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell 1, 346–352 (2007).
Surti, U., Hoffner, L., Chakravarti, A. & Ferrell, R.E. Genetics and biology of human ovarian teratomas. I. Cytogenetic analysis and mechanism of origin. Am. J. Hum. Genet. 47, 635–643 (1990).
Pick, M. et al. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells 27, 2686–2690 (2009).
Luedi, P.P. et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 17, 1723–1730 (2007).
Stadtfeld, M. et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181 (2010).
Tucker, K.L. et al. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10, 1008–1020 (1996).
Sontheimer, E.J. & Steitz, J.A. Three novel functional variants of human U5 small nuclear RNA. Mol. Cell. Biol. 12, 734–746 (1992).
Edwards, C.A. & Ferguson-Smith, A.C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 19, 281–289 (2007).
Chabot, B., Black, D.L., LeMaster, D.M. & Steitz, J.A. The 3′ splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science 230, 1344–1349 (1985).
Patterson, B. & Guthrie, C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell 49, 613–624 (1987).
Hentze, M.W. & Kulozik, A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96, 307–310 (1999).
Seitz, H. et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261–262 (2003).
Seitz, H. et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 14, 1741–1748 (2004).
Suh, M.R. et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270, 488–498 (2004).
Voorhoeve, P.M. et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181 (2006).
Fundele, R.H. et al. Temporal and spatial selection against parthenogenetic cells during development of fetal chimeras. Development 108, 203–211 (1990).
Thomson, J.A. & Solter, D. The developmental fate of androgenetic, parthenogenetic, and gynogenetic cells in chimeric gastrulating mouse embryos. Genes Dev. 2, 1344–1351 (1988).
Thomson, J.A. & Solter, D. Chimeras between parthenogenetic or androgenetic blastomeres and normal embryos: allocation to the inner cell mass and trophectoderm. Dev. Biol. 131, 580–583 (1989).
Nagy, A., Sass, M. & Markkula, M. Systematic non-uniform distribution of parthenogenetic cells in adult mouse chimaeras. Development 106, 321–324 (1989).
Strain, L., Warner, J.P., Johnston, T. & Bonthron, D.T. A human parthenogenetic chimaera. Nat. Genet. 11, 164–169 (1995).
Xu, R.H. et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 (2002).
Will, C.L. & Luhrmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290–301 (2001).
Karijolich, J. & Yu, Y.T. Spliceosomal snRNA modifications and their function. RNA Biol. 7, 192–204 (2010).
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21–32 (2001).
Murchison, E.P., Partridge, J.F., Tam, O.H., Cheloufi, S. & Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 102, 12135–12140 (2005).
Kanellopoulou, C. et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 (2005).
Park, I.H., Lerou, P.H., Zhao, R., Huo, H. & Daley, G.Q. Generation of human induced pluripotent stem cells. Nat. Protoc. 3, 1180–1186 (2008).
Le Bibikova, M. et al. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics 1, 177–200 (2009).
Acknowledgements
We thank M. Pick for her help with generating parthenogenetic iPSCs and O. Bar-Nur for providing WT-HiPSCs; U. Ben-David for his technical help with the teratoma formation experiments; T. Golan-Lev for her assistance with the graphic design and immunostaining assays; and A. Eden, D. Kitsberg, Y. Mayshar, D. Ronen and N. Sharon for critical reading of the manuscript. N.B. is the Herbert Cohn Chair in Cancer Research. This research was supported partially by the Israel Science Foundation–Morasha Foundation (grant no. 943/09).
Author information
Authors and Affiliations
Contributions
Y.S.: conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; O.Y.: conception and design, and collection and assembly of data; N.B.: conception and design, financial support, data analysis and interpretation, manuscript writing and final approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures 1–8
Supplementary Figures 1–8 (PDF 7072 kb)
Supplementary Table 1
Expression profile of differentially expressed genes and mirRNAs (XLS 435 kb)
Supplementary Table 2
Tissue specific gene expression analysis (XLS 568 kb)
Supplementary Table 3
Gene expression analysis following 7 days of treatment with BMP4 (XLS 25 kb)
Supplementary Table 4
Full list of primers (DOC 39 kb)
Rights and permissions
About this article
Cite this article
Stelzer, Y., Yanuka, O. & Benvenisty, N. Global analysis of parental imprinting in human parthenogenetic induced pluripotent stem cells. Nat Struct Mol Biol 18, 735–741 (2011). https://doi.org/10.1038/nsmb.2050
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2050
This article is cited by
-
High levels of unfolded protein response component CHAC1 associates with cancer progression signatures in malignant breast cancer tissues
Clinical and Translational Oncology (2022)
-
Generation of human androgenetic induced pluripotent stem cells
Scientific Reports (2020)
-
Higher expression of cation transport regulator-like protein 1 (CHAC1) predicts of poor outcomes in uveal melanoma (UM) patients
International Ophthalmology (2019)
-
Genotypes of cancer stem cells characterized by epithelial-to-mesenchymal transition and proliferation related functions
Scientific Reports (2016)
-
Pluripotent stem cells in disease modelling and drug discovery
Nature Reviews Molecular Cell Biology (2016)