Key Points
-
Large-scale genome-wide association studies (GWAS), meta-analyses, fine-mapping and studies with populations of diverse ancestries have identified hundreds of genetic loci that predispose individuals to rheumatic diseases
-
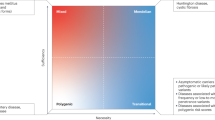
Genetic susceptibility regions overlap between diseases, suggesting common disease mechanisms and treatment choices
-
Genetic associations can implicate pathways (for example, T helper cell differentiation) involved in disease onset and/or outcome
-
A number of existing drugs (such as tofacitinib, tocilizumab and abatacept) target proteins encoded by genes identified through GWAS, suggesting that genetic studies can help identify novel therapeutic targets
-
Most disease-associated variants map to noncoding regions responsible for controlling gene expression and DNA conformational analysis is beginning to help in the challenging task of unravelling their function
Abstract
Susceptibility to rheumatic diseases, such as osteoarthritis, rheumatoid arthritis, ankylosing spondylitis, systemic lupus erythematosus, juvenile idiopathic arthritis and psoriatic arthritis, includes a large genetic component. Understanding how an individual's genetic background influences disease onset and outcome can lead to a better understanding of disease biology, improved diagnosis and treatment, and, ultimately, to disease prevention or cure. The past decade has seen great progress in the identification of genetic variants that influence the risk of rheumatic diseases. The challenging task of unravelling the function of these variants is ongoing. In this Review, the major insights from genetic studies, gained from advances in technology, bioinformatics and study design, are discussed in the context of rheumatic disease. In addition, pivotal genetic studies in the main rheumatic diseases are highlighted, with insights into how these studies have changed the way we view these conditions in terms of disease overlap, pathways of disease and potential new therapeutic targets. Finally, the limitations of genetic studies, gaps in our knowledge and ways in which current genetic knowledge can be fully translated into clinical benefit are examined.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Alarcon-Segovia, D. et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 52, 1138–1147 (2005).
Brown, M. A. et al. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum. 40, 1823–1828 (1997).
Karason, A., Love, T. J. & Gudbjornsson, B. A strong heritability of psoriatic arthritis over four generations — the Reykjavik Psoriatic Arthritis Study. Rheumatology (Oxford) 48, 1424–1428 (2009).
Silman, A. J. et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br. J. Rheumatol. 32, 903–907 (1993).
Spector, T. D. & MacGregor, A. J. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage 12 (Suppl. A), S39–S44 (2004).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Plenge, R. M. et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet. 39, 1477–1482 (2007).
Plenge, R. M. et al. TRAF1-C5 as a risk locus for rheumatoid arthritis — a genomewide study. N. Engl. J. Med. 357, 1199–1209 (2007).
Remmers, E. F. et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N. Engl. J. Med. 357, 977–986 (2007).
Barton, A. et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat. Genet. 40, 1156–1159 (2008).
Raychaudhuri, S. et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 41, 1313–1318 (2009).
Stahl, E. A. et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514 (2010).
Thomson, W. et al. Rheumatoid arthritis association at 6q23. Nat. Genet. 39, 1431–1433 (2007).
Burton, P. R. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Evans, D. M. et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 (2011).
Reveille, J. D. et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet. 42, 123–127 (2010).
Zeggini, E. et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 380, 815–823 (2012).
Valdes, A. M. et al. Involvement of different risk factors in clinically severe large joint osteoarthritis according to the presence of hand interphalangeal nodes. Arthritis Rheum. 62, 2688–2695 (2010).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Evangelou, E. et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann. Rheum. Dis. 73, 2130–2136 (2014).
Gee, F., Rushton, M. D., Loughlin, J. & Reynard, L. N. Correlation of the osteoarthritis susceptibility variants that map to chromosome 20q13 with an expression quantitative trait locus operating on NCOA3 and with functional variation at the polymorphism rs116855380. Arthritis Rheumatol. 67, 2923–2932 (2015).
Bentham, J. et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 47, 1457–1464 (2015).
Morris, D. L. et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 48, 940–946 (2016).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Chapman, K. et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5′ UTR of GDF5 with osteoarthritis susceptibility. Hum. Mol. Genet. 17, 1497–1504 (2008).
Ellinghaus, D. et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48, 510–518 (2016).
Adrianto, I. et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 43, 253–258 (2011).
Eyre, S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012).
Cortes, A. et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 (2013).
Hinks, A. et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 45, 664–669 (2013).
Sun, C. et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 48, 323–330 (2016).
Fortune, M. D. et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat. Genet. 47, 839–846 (2015).
Liley, J. & Wallace, C. A pleiotropy-informed Bayesian false discovery rate adapted to a shared control design finds new disease associations from GWAS summary statistics. PLoS Genet. 11, e1004926 (2015).
Wallace, C. et al. Dissection of a complex disease susceptibility region using a Bayesian stochastic search approach to fine mapping. PLoS Genet. 11, e1005272 (2015).
Bowes, J. et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat. Commun. 6, 6046 (2015).
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012).
Gaffen, S. L., Jain, R., Garg, A. V. & Cua, D. J. The IL-23–IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 14, 585–600 (2014).
Onengut-Gumuscu, S. et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 47, 381–386 (2015).
Lemos, L. L. et al. Treatment of psoriatic arthritis with anti-TNF agents: a systematic review and meta-analysis of efficacy, effectiveness and safety. Rheumatol. Int. 34, 1345–1360 (2014).
Bossini-Castillo, L., Lopez-Isac, E. & Martin, J. Immunogenetics of systemic sclerosis: defining heritability, functional variants and shared-autoimmunity pathways. J. Autoimmun. 64, 53–65 (2015).
Mayes, M. D. et al. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am. J. Hum. Genet. 94, 47–61 (2014).
Lopez-Isac, E. et al. Brief report: IRF4 newly identified as a common susceptibility locus for systemic sclerosis and rheumatoid arthritis in a cross-disease meta-analysis of genome-wide association studies. Arthritis Rheumatol. 68, 2338–2344 (2016).
Sekine, C. et al. Successful treatment of animal models of rheumatoid arthritis with small-molecule cyclin-dependent kinase inhibitors. J. Immunol. 180, 1954–1961 (2008).
Ferreira, M. A. et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378, 1006–1014 (2011).
Begovich, A. B. et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 75, 330–337 (2004).
Jostins, L. et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Mohan, C. & Putterman, C. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat. Rev. Nephrol. 11, 329–341 (2015).
Guerra, S. G., Vyse, T. J. & Cunninghame Graham, D. S. The genetics of lupus: a functional perspective. Arthritis Res. Ther. 14, 211 (2012).
Robinson, P. C. & Brown, M. A. Genetics of ankylosing spondylitis. Mol. Immunol. 57, 2–11 (2014).
Brown, M. A., Kenna, T. & Wordsworth, B. P. Genetics of ankylosing spondylitis — insights into pathogenesis. Nat. Rev. Rheumatol. 12, 81–91 (2016).
Parkes, M., Cortes, A., van Heel, D. A. & Brown, M. A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14, 661–673 (2013).
Loughlin, J. Genetic contribution to osteoarthritis development: current state of evidence. Curr. Opin. Rheumatol. 27, 284–288 (2015).
Reynard, L. N. & Loughlin, J. Insights from human genetic studies into the pathways involved in osteoarthritis. Nat. Rev. Rheumatol. 9, 573–583 (2013).
Rogers, E. L., Reynard, L. N. & Loughlin, J. The role of inflammation-related genes in osteoarthritis. Osteoarthritis Cartilage 23, 1933–1938 (2015).
Lessard, C. J. et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren's syndrome. Nat. Genet. 45, 1284–1292 (2013).
Li, Y. et al. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjogren's syndrome at 7q11.23. Nat. Genet. 45, 1361–1365 (2013).
Albagha, O. M. et al. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget's disease of bone. Nat. Genet. 42, 520–524 (2010).
Albagha, O. M. et al. Genome-wide association identifies three new susceptibility loci for Paget's disease of bone. Nat. Genet. 43, 685–689 (2011).
Vallet, M. et al. Targeted sequencing of the Paget's disease associated 14q32 locus identifies several missense coding variants in RIN3 that predispose to Paget's disease of bone. Hum. Mol. Genet. 24, 3286–3295 (2015).
Lyons, P. A. et al. Genetically distinct subsets within ANCA-associated vasculitis. N. Engl. J. Med. 367, 214–223 (2012).
Xie, G. et al. Association of granulomatosis with polyangiitis (Wegener's) with HLA-DPB1*04 and SEMA6A gene variants: evidence from genome-wide analysis. Arthritis Rheum. 65, 2457–2468 (2013).
Rahmattulla, C. et al. Genetic variants in ANCA-associated vasculitis: a meta-analysis. Ann. Rheum. Dis. 75, 1687–1692 (2016).
Kottgen, A. et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet. 45, 145–154 (2013).
Miao, Z. M. et al. NALP3 inflammasome functional polymorphisms and gout susceptibility. Cell Cycle 8, 27–30 (2009).
Detert, J. & Klaus, P. Biologic monotherapy in the treatment of rheumatoid arthritis. Biologics 9, 35–43 (2015).
Shetty, A. et al. Tocilizumab in the treatment of rheumatoid arthritis and beyond. Drug Des. Devel. Ther. 8, 349–364 (2014).
Ferreira, R. C. et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 9, e1003444 (2013).
Swerdlow, D. I. et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet 379, 1214–1224 (2012).
Schiff, M. Abatacept treatment for rheumatoid arthritis. Rheumatology (Oxford) 50, 437–449 (2011).
Oliver, J., Plant, D., Webster, A. P. & Barton, A. Genetic and genomic markers of anti-TNF treatment response in rheumatoid arthritis. Biomark. Med. 9, 499–512 (2015).
Plant, D., Wilson, A. G. & Barton, A. Genetic and epigenetic predictors of responsiveness to treatment in RA. Nat. Rev. Rheumatol. 10, 329–337 (2014).
Farh, K. K. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Bernstein, B. E. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010).
Kersey, P. J. et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44, D574–D580 (2016).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Martens, J. H. & Stunnenberg, H. G. BLUEPRINT: mapping human blood cell epigenomes. Haematologica 98, 1487–1489 (2013).
Yates, A. et al. Ensembl 2016. Nucleic Acids Res. 44, D710–D716 (2016).
Trynka, G. & Raychaudhuri, S. Using chromatin marks to interpret and localize genetic associations to complex human traits and diseases. Curr. Opin. Genet. Dev. 23, 635–641 (2013).
GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Carithers, L. J. & Moore, H. M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank. 13, 307–308 (2015).
Fairfax, B. P. & Knight, J. C. Genetics of gene expression in immunity to infection. Curr. Opin. Immunol. 30, 63–71 (2014).
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014).
Westra, H. J. et al. Cell specific eQTL analysis without sorting cells. PLoS Genet. 11, e1005223 (2015).
Andres, A. M. et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 6, e1001157 (2010).
Robinson, P. C. et al. ERAP2 is associated with ankylosing spondylitis in HLA-B27-positive and HLA-B27-negative patients. Ann. Rheum. Dis. 74, 1627–1629 (2015).
Graham, R. R. et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl Acad. Sci. USA 104, 6758–6763 (2007).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Belton, J. M. et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Miele, A. & Dekker, J. Mapping cis- and trans- chromatin interaction networks using chromosome conformation capture (3C). Methods Mol. Biol. 464, 105–121 (2009).
Martin, P. et al. Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nat. Commun. 6, 10069 (2015).
Grabiec, A. M. et al. JNK-dependent downregulation of FoxO1 is required to promote the survival of fibroblast-like synoviocytes in rheumatoid arthritis. Ann. Rheum. Dis. 74, 1763–1771 (2015).
Nakano, K., Whitaker, J. W., Boyle, D. L., Wang, W. & Firestein, G. S. DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis. 72, 110–117 (2013).
Wang, S., Wen, F., Wiley, G. B., Kinter, M. T. & Gaffney, P. M. An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression. PLoS Genet. 9, e1003750 (2013).
McGovern, A. et al. Capture Hi-C identifies a novel causal gene, IL20RA, in the pan-autoimmune genetic susceptibility region 6q23. Genome Biol. 17, 212 (2016).
Cong, L. & Zhang, F. Genome engineering using CRISPR–Cas9 system. Methods Mol. Biol. 1239, 197–217 (2015).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Wright, A. V., Nunez, J. K. & Doudna, J. A. Biology and applications of CRISPRsystems: harnessing nature's toolbox for genome engineering. Cell 164, 29–44 (2016).
Hilton, I. B. & Gersbach, C. A. Enabling functional genomics with genome engineering. Genome Res. 25, 1442–1455 (2015).
Hilton, I. B. et al. Epigenome editing by a CRISPR–Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015).
Thakore, P. I. et al. Highly specific epigenome editing by CRISPR–Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015).
Thakore, P. I., Black, J. B., Hilton, I. B. & Gersbach, C. A. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods 13, 127–137 (2016).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
The International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
Zhernakova, A. et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 7, e1002004 (2011).
McCarthy, M. I. et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369 (2008).
Evangelou, E. & Ioannidis, J. P. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 14, 379–389 (2013).
Li, Y. R. & Keating, B. J. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 6, 91 (2014).
Konig, I. R. Validation in genetic association studies. Brief. Bioinform. 12, 253–258 (2011).
Stephens, M. & Balding, D. J. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet. 10, 681–690 (2009).
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR–Cas9. Science 346, 1258096 (2014).
Albert, F. W. & Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16, 197–212 (2015).
Ioannidis, J. P., Thomas, G. & Daly, M. J. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 10, 318–329 (2009).
Nejentsev, S., Walker, N., Riches, D., Egholm, M. & Todd, J. A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324, 387–389 (2009).
Pe'er, I. et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat. Genet. 38, 663–667 (2006).
Lewis-Faning, E. Report on an enquiry into the aetiological factors associated with rheumatoid arthritis. Ann. Rheum. Dis. 9, 1–94 (1950).
Stastny, P. Mixed lymphocyte cultures in rheumatoid arthritis. J. Clin. Invest. 57, 1148–1157 (1976).
Gregersen, P. K., Silver, J. & Winchester, R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 30, 1205–1213 (1987).
MacGregor, A. J. et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 43, 30–37 (2000).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Klein, R. J. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005).
Abecasis, G. R. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Acknowledgements
The authors' research work was supported by the Wellcome Trust (grant number 095684), Arthritis Research UK (grant number 20385) and the National Institute for Health Research Manchester Musculoskeletal Biomedical Research Unit. The views expressed in this publication are those of the authors and not necessarily those of the UK National Health Service, the National Institute for Health Research or the UK Department of Health.
Author information
Authors and Affiliations
Contributions
All authors researched the data for the article, provided substantial contributions to discussions of its content, wrote the article and undertook review and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Single nucleotide polymorphism (SNP)
-
DNA sequence variation of a single nucleotide that occurs at a specific position in the genome.
- Candidate gene
-
In genetic association studies, the candidate gene approach tests associations between genetic variation within pre-specified genes of interest and phenotypes.
- Genome-wide significance threshold
-
P value threshold that is considered statistically significant after multiple testing correction in GWAS, typically P <5 × 10−8.
- Non-synonymous SNPs
-
SNPs that map to coding genes and result in an amino acid change in the protein they encode.
- Statistical power
-
Probability that a test correctly rejects the null hypothesis.
- Risk score
-
Estimate of the cumulative contribution of genetic factors to a specific outcome of interest in an individual that takes into account the reported risk alleles.
- Linkage disequilibrium
-
Nonrandom association of alleles at different loci in a given population.
Rights and permissions
About this article
Cite this article
Eyre, S., Orozco, G. & Worthington, J. The genetics revolution in rheumatology: large scale genomic arrays and genetic mapping. Nat Rev Rheumatol 13, 421–432 (2017). https://doi.org/10.1038/nrrheum.2017.80
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2017.80
This article is cited by
-
A Common Functional Variant at the Enhancer of the Rheumatoid Arthritis Risk Gene ORMDL3 Regulates its Expression Through Allele-Specific JunD Binding
Phenomics (2023)
-
Associations between adipokines gene polymorphisms and knee osteoarthritis: a meta-analysis
BMC Musculoskeletal Disorders (2022)
-
How does age determine the development of human immune-mediated arthritis?
Nature Reviews Rheumatology (2022)
-
An introduction to machine learning and analysis of its use in rheumatic diseases
Nature Reviews Rheumatology (2021)
-
Rheumatological manifestations in inborn errors of immunity
Pediatric Research (2020)