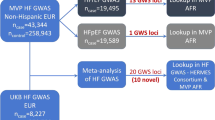
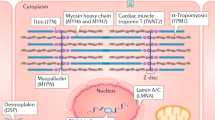
Abstract
Heart failure is an increasingly prevalent and highly lethal disease that is most often caused by underlying pathologies, such as myocardial infarction or hypertension, but it can also be the result of a single gene mutation. Comprehensive genetic and genomic approaches are starting to disentangle the diverse molecular underpinnings of both forms of the disease and promise to yield much-needed novel diagnostic and therapeutic options for specific subtypes of heart failure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Hunt, S. A. et al. 2009 focused update incorporated into the ACC/AHA 2005 guidelines for the diagnosis and management of heart failure in adults: a report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines: developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation 119, e391–e479 (2009).
Creemers, E. E. & Pinto, Y. M. Molecular mechanisms that control interstitial fibrosis in the pressure-overloaded heart. Cardiovasc. Res. 89, 265–272 (2010).
Swynghedauw, B. Molecular mechanisms of myocardial remodeling. Physiol. Rev. 79, 215–262 (1999).
Benjamin, I. J. & Schneider, M. D. Learning from failure: congestive heart failure in the postgenomic age. J. Clin. Invest. 115, 495–499 (2005).
Mudd, J. O. & Kass, D. A. Tackling heart failure in the twenty-first century. Nature 451, 919–928 (2008).
Lee, D. S. et al. Association of parental heart failure with risk of heart failure in offspring. N. Engl. J. Med. 355, 138–147 (2006).
Smith, N. L. et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ. Cardiovasc. Genet. 3, 256–266 (2010).
Morrison, A. C. et al. Genomic variation associated with mortality among adults of European and african ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ. Cardiovasc. Genet. 3, 248–255 (2010).
Vasan, R. S. et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA 302, 168–178 (2009).
Cappola, T. P. et al. Loss-of-function DNA sequence variant in the CLCNKA chloride channel implicates the cardio-renal axis in interindividual heart failure risk variation. Proc. Natl Acad. Sci. USA 108, 2456–2461 (2011).
Hershberger, R. E. et al. Genetic evaluation of cardiomyopathy-a Heart Failure Society of America practice guideline. J. Card. Fail. 15, 83–97 (2009).
van der Zwaag, P. A. et al. One mutation fits all: phospolamban R14del causes both dilated cardiomypathy and arrhytmogenic right ventricular cardiomyopathy/dysplasia. Circulation 122, A17663 (2010).
van Spaendonck-Zwarts, K. Y. et al. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation 121, 2169–2175 (2010).
Ng, S. B. et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature Genet. 42, 790–793 (2010).
Zhang, C. L. et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 (2002).
Movassagh, M. et al. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS ONE 5, e8564 (2010).
Yi, P., Han, Z., Li, X. & Olson, E. N. The mevalonate pathway controls heart formation in Drosophila by isoprenylation of Gγ1. Science 313, 1301–1303 (2006).
Milan, D. J. et al. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation 120, 553–559 (2009).
Lowes, B. D. et al. Myocardial gene expression in dilated cardiomyopathy treated with β-blocking agents. N. Engl. J. Med. 346, 1357–1365 (2002).
Knollmann, B. C. & Roden, D. M. A genetic framework for improving arrhythmia therapy. Nature 451, 929–936 (2008).
Nanni, L., Romualdi, C., Maseri, A. & Lanfranchi, G. Differential gene expression profiling in genetic and multifactorial cardiovascular diseases. J. Mol. Cell. Cardiol. 41, 934–948 (2006).
Hwang, J. J. et al. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol. Genomics 10, 31–44 (2002).
Schroen, B. et al. Thrombospondin-2 is essential for myocardial matrix integrity: increased expression identifies failure-prone cardiac hypertrophy. Circ. Res. 95, 515–522 (2004).
Schroen, B. et al. Lysosomal integral membrane protein 2 is a novel component of the cardiac intercalated disc and vital for load-induced cardiac myocyte hypertrophy. J. Exp. Med. 204, 1227–1235 (2007).
van Kimmenade, R. R. et al. Utility of amino-terminal pro-brain natriuretic peptide, galectin-3, and apelin for the evaluation of patients with acute heart failure. J. Am. Coll. Cardiol. 48, 1217–1224 (2006).
Matkovich, S. J., Zhang, Y., Van Booven, D. J. & Dorn, G. W. Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Gαq. Circ. Res. 106, 1459–1467 (2010).
Creemers, E. E., Sutherland, L. B., Oh, J., Barbosa, A. C. & Olson, E. N. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol. Cell 23, 83–96 (2006).
Kong, S. W. et al. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ. Cardiovasc. Genet. 3, 138–146 (2010).
Neagoe, C. et al. Titin isoform switch in ischemic human heart disease. Circulation 106, 1333–1341 (2002).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotech. 28, 511–515 (2010).
Kleinjan, D. A. & van Heyningen, V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32 (2005).
Blow, M. J. et al. ChIP-Seq identification of weakly conserved heart enhancers. Nature Genet. 42, 806–810 (2010).
Haberland, M., Montgomery, R. L. & Olson, E. N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Rev. Genet. 10, 32–42 (2009).
Rakyan, V. K. & Beck, S. Epigenetic variation and inheritance in mammals. Curr. Opin. Genet. Dev. 16, 573–577 (2006).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Thum, T. et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 (2008).
Care, A. et al. MicroRNA-133 controls cardiac hypertrophy. Nature Med. 13, 613–618 (2007).
Small, E. M., Frost, R. J. & Olson, E. N. MicroRNAs add a new dimension to cardiovascular disease. Circulation 121, 1022–1032 (2010).
van Rooij, E. et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007).
Elmen, J. et al. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008).
Tijsen, A. J. et al. MiR423–425p as a circulating biomarker for heart failure. Circ. Res. 106, 1035–1039 (2010).
Clop, A. et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nature Genet. 38, 813–818 (2006).
Kasowski, M. et al. Variation in transcription factor binding among humans. Science 328, 232–235 (2010).
Dorn, G. W. Pharmacogenetic profiling in the treatment of heart disease. Transl. Res. 154, 295–302 (2009).
Johnson, J. A. & Liggett, S. B. Cardiovascular pharmacogenomics of adrenergic receptor signaling: clinical implications and future directions. Clin. Pharmacol. Ther. 89, 366–378 (2011).
Brugts, J. J. et al. Genetic determinants of treatment benefit of the angiotensin-converting enzyme-inhibitor perindopril in patients with stable coronary artery disease. Eur. Heart J. 31, 1854–1864 (2010).
Sakuntabhai, A. et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nature Genet. 21, 271–277 (1999).
Mayosi, B. M. et al. Heterozygous disruption of SERCA2a is not associated with impairment of cardiac performance in humans: implications for SERCA2a as a therapeutic target in heart failure. Heart 92, 105–109 (2006).
Shull, G. E. et al. Physiological functions of plasma membrane and intracellular Ca2+ pumps revealed by analysis of null mutants. Ann. N. Y. Acad. Sci. 986, 453–460 (2003).
Acknowledgements
The authors are grateful to M. van den Hoogenhof for his help in preparing this manuscript and B. Knollmann for critical reading of this manuscript. The authors acknowledge grant support from the Center of Translational Molecular Medicine (CTMM), the European Commission project INHERITANCE, The InterUniversity Cardiology Institute Netherlands (ICIN) and the Netherlands Heart Foundation (NHS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Y.M.P. has a minor interest in a spin-off company that has rights relating to the biomarker galectin 3, which is mentioned in this article.
Supplementary information
Supplementary information S1 (table)
Genes linked to Mendelian forms of heart failure. (PDF 265 kb)
Related links
Glossary
- Interstitial fibrosis
-
Excessive deposition and accumulation of extracellular matrix proteins in the cardiac interstitium.
- Kabuki syndrome
-
A rare congenital disorder, named after the facial resemblance of affected individuals to white Kabuki makeup, a Japanese traditional theatrical form. There is a wide range of congenital problems associated with Kabuki syndrome, including heart defects, urinary tract anomalies, hearing loss and postnatal growth deficiency.
- Odds ratio
-
The ratio of the odds of disease between two groups, in which the odds describe the probability of being affected divided by the probability of not being affected.
- Peri-partum cardiomyopathy
-
A rare but often fatal form of dilated cardiomyopathy that is defined as heart failure occurring in the mother in the last months of pregnancy or in the first 6 months after delivery. It occurs in the absence of an identifiable cause, and without demonstrable pre-existing or concurrent heart disease.
- Sarcomere
-
The basic contractile unit of a muscle cell.
- Tachyarrhythmias or bradyarrhythmias
-
An excessively rapid or excessively slow heartbeat of more than 120 or less than 50 beats per minute, respectively.
Rights and permissions
About this article
Cite this article
Creemers, E., Wilde, A. & Pinto, Y. Heart failure: advances through genomics. Nat Rev Genet 12, 357–362 (2011). https://doi.org/10.1038/nrg2983
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2983
This article is cited by
-
Dilated cardiomyopathy in the era of precision medicine: latest concepts and developments
Heart Failure Reviews (2022)
-
A personalized, multiomics approach identifies genes involved in cardiac hypertrophy and heart failure
npj Systems Biology and Applications (2018)
-
Cardiac molecular imaging to track left ventricular remodeling in heart failure
Journal of Nuclear Cardiology (2017)
-
HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy
Molecular and Cellular Biochemistry (2017)
-
Genetic Predispositions to Heart Failure
Current Cardiovascular Risk Reports (2016)