Abstract
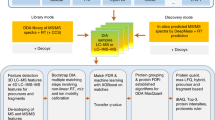
Identifying the species on which hematophagous arthropods feed is crucial for studying the factors that affect pathogen distributions and that can aid public health. Here we describe a protocol to identify the species a parasitic arthropod has previously fed upon by identifying the source of the remnants of a previous blood meal via shotgun proteomics and spectral matching. The protocol is a nontargeted approach that uses the entire detected blood proteome for source identification; it does not require a priori knowledge of genome or protein sequences. Instead, reference spectral libraries are compiled from the blood of multiple host species by using SpectraST, which takes ∼4 d; the identification of the species from which a previous blood meal of a hematophagous arthropod was taken is achieved with spectral matching against the reference spectral libraries, which takes approximately another 4 d. This method is robust against random degradation of the blood meal and can identify unknown blood remnants months after the feeding event.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Taylor, L.H., Latham, S.M. & Woolhouse, M.E. Risk factors for human disease emergence. Philos. Trans. Royal Soc. Lond. B Biol. Sci. 356, 983–989 (2001).
Leibold, M.A. A graphical model of keystone predators in food webs: trophic regulation of abundance, incidence, and diversity patterns in communities. Am. Naturalist 147, 784–812 (1996).
Rose, M.D. & Polis, G.A. The distribution and abundance of coyotes: the effects of allochthonous food subsidies from the sea. Ecology 79, 998–1007 (1998).
LoGiudice, K., Ostfeld, R.S., Schmidt, K.A. & Keesing, F. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc. Natl. Acad. Sci. USA 100, 567–571 (2003).
Lord, R.D., Lord, V.R., Humphreys, J.G. & McLean, R.G. Distribution of Borrelia burgdorferi in host mice in Pennsylvania. J. Clin. Microbiol. 32, 2501–2504 (1994).
Matuschka, F.R., Fischer, P., Musgrave, K., Richter, D. & Spielman, A. Hosts on which nymphal Ixodes ricinus most abundantly feed. Am. J. Trop. Med. Hyg. 44, 100–107 (1991).
Brisson, D., Dykhuizen, D.E. & Ostfeld, R.S. Conspicuous impacts of inconspicuous hosts on the Lyme disease epidemic. Proc. Roy. Soc. Biol. Sci. 275, 227–235 (2008).
Allan, B.F., Goessling, L.S., Storch, G.A. & Thach, R.E. Blood meal analysis to identify reservoir hosts for Amblyomma americanum ticks. Emerg. Infect. Dis. 16, 433–440 (2010).
Arnold, E.H., Simmons, S.W. & Fawcett, D.G. Precipitin technique for determining mosquito blood meals. Public Health Rep. 61, 1244–1249 (1946).
Burkot, T.R., Goodman, W.G. & DeFoliart, G.R. Identification of mosquito blood meals by enzyme-linked immunosorbent assay. Am. J. Trop. Med. Hyg. 30, 1336–1341 (1981).
Kent, R.J. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol. Ecol. Resour. 9, 4–18 (2009).
Tempelis, C.H. & Rodrick, M.L. Passive hemagglutination inhibition technique for the identification of arthropod blood meals. Am. J. Trop. Med. Hyg. 21, 238–245 (1972).
Wickramasekara, S., Bunikis, J., Wysocki, V. & Barbour, A.G. Identification of residual blood proteins in ticks by mass spectrometry proteomics. Emerg. Infect. Dis. 14, 1273–1275 (2008).
Schubert, J.H. & Holdeman, L.V. A modified precipitin technique for determining the source of mosquito blood-meals. Am. J. Trop. Med. Hyg. 5, 272–273 (1956).
Onder, O., Shao, W., Kemps, B., Lam, H. & Brisson, D. Blood meal source tracking through genome-free proteomics technology using unidentified tandem mass spectral libraries. Nat. Commun. 4, 1746 (2013).
Laskay, U.A. et al. Development of a host blood meal database: de novo sequencing of hemoglobin from nine small mammals using mass spectrometry. Biol. Chem. 393, 195–201 (2012).
Gariepy, T.D., Lindsay, R., Ogden, N. & Gregory, T.R. Identifying the last supper: utility of the DNA barcode library for bloodmeal identification in ticks. Mol. Ecol. Resour. 12, 646–652 (2012).
Mukabana, W.R., Takken, W. & Knols, B.G. Analysis of arthropod bloodmeals using molecular genetic markers. Trends Parasitol. 18, 505–509 (2002).
Humair, P.F. et al. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J. Med. Entomol. 44, 869–880 (2007).
Pizarro, J.C. & Stevens, L. A new method for forensic DNA analysis of the blood meal in Chagas disease vectors demonstrated using Triatoma infestans from Chuquisaca, Bolivia. PLoS ONE 3, e3585 (2008).
Mota, J. et al. Identification of blood meal source and infection with Trypanosoma cruzi of Chagas disease vectors using a multiplex cytochrome b polymerase chain reaction assay. Vector Borne Zoonotic Dis. 7, 617–627 (2007).
Moran Cadenas, F. et al. Identification of host bloodmeal source and Borrelia burgdorferi sensu lato in field-collected Ixodes ricinus ticks in Chaumont (Switzerland). J. Med. Entomol. 44, 1109–1117 (2007).
Dasari, S. et al. Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment. J. Proteome Res. 11, 1686–1695 (2012).
Lam, H. & Aebersold, R. Building and searching tandem mass (MS/MS) spectral libraries for peptide identification in proteomics. Methods 54, 424–431 (2011).
Lam, H. et al. Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 (2008).
Craig, R., Cortens, J.C., Fenyo, D. & Beavis, R.C. Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 5, 1843–1849 (2006).
Frewen, B.E., Merrihew, G.E., Wu, C.C., Noble, W.S. & MacCoss, M.J. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 78, 5678–5684 (2006).
Stein, S.E. & Scott, D.R. Optimization and testing of mass-spectral library search algorithms for compound identification. J. Am. Soc. Mass Spectr. 5, 859–866 (1994).
Onder, O., Turkarslan, S., Sun, D. & Daldal, F. Overproduction or absence of the periplasmic protease DegP severely compromises bacterial growth in the absence of the dithiol: disulfide oxidoreductase DsbA. Mol. Cell Proteomics 7, 875–890 (2008).
Onder, O. et al. Modifications of the lipoamide-containing mitochondrial subproteome in a yeast mutant defective in cysteine desulfurase. Mol. Cell Proteomics 5, 1426–1436 (2006).
Chambers, M.C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Acknowledgements
This work was supported by a grant from the US National Institutes of Health (AI076342); the US Center for Disease Control and Prevention (U01CK000170); and the University Grant Council, Hong Kong Special Administrative Region Government (HKUST RPC10EG08, DAG12EG01S).
Author information
Authors and Affiliations
Contributions
D.B., H.L. and Ö.Ö. conceived the study and formulated the experimental and computational strategy. Ö.Ö. optimized all the experimental protocols. H.L. and W.S. adapted SpectraST to build spectral libraries from unidentified spectra. Ö.Ö., H.L. and D.B. co-wrote the manuscript, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Önder, Ö., Shao, W., Lam, H. et al. Tracking the sources of blood meals of parasitic arthropods using shotgun proteomics and unidentified tandem mass spectral libraries. Nat Protoc 9, 842–850 (2014). https://doi.org/10.1038/nprot.2014.048
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.048
This article is cited by
-
Blood meal acquisition enhances arbovirus replication in mosquitoes through activation of the GABAergic system
Nature Communications (2017)
-
How do biting disease vectors behaviourally respond to host availability?
Parasites & Vectors (2016)
-
Reliability of molecular host-identification methods for ticks: an experimental in vitro study with Ixodes ricinus
Parasites & Vectors (2015)
-
Top-Down-Assisted Bottom-Up Method for Homologous Protein Sequencing: Hemoglobin from 33 Bird Species
Journal of the American Society for Mass Spectrometry (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.