Abstract
Immune responses are tightly regulated to ensure efficient pathogen clearance while avoiding tissue damage. Here we report that Setdb2 was the only protein lysine methyltransferase induced during infection with influenza virus. Setdb2 expression depended on signaling via type I interferons, and Setdb2 repressed expression of the gene encoding the neutrophil attractant CXCL1 and other genes that are targets of the transcription factor NF-κB. This coincided with occupancy by Setdb2 at the Cxcl1 promoter, which in the absence of Setdb2 displayed diminished trimethylation of histone H3 Lys9 (H3K9me3). Mice with a hypomorphic gene-trap construct of Setdb2 exhibited increased infiltration of neutrophils during sterile lung inflammation and were less sensitive to bacterial superinfection after infection with influenza virus. This suggested that a Setdb2-mediated regulatory crosstalk between the type I interferons and NF-κB pathways represents an important mechanism for virus-induced susceptibility to bacterial superinfection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Morens, D.M., Taubenberger, J.K. & Fauci, A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970 (2008).
Molinari, N.A. et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25, 5086–5096 (2007).
van der Sluijs, K.F., van der Poll, T., Lutter, R., Juffermans, N.P. & Schultz, M.J. Bench-to-bedside review: bacterial pneumonia with influenza - pathogenesis and clinical implications. Crit. Care 14, 219 (2010).
McCullers, J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 12, 252–262 (2014).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 (2001).
Sadler, A.J. & Williams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 (2008).
Schoggins, J.W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014).
Oeckinghaus, A. & Ghosh, S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 1, a000034 (2009).
Medzhitov, R., Schneider, D.S. & Soares, M.P. Disease tolerance as a defense strategy. Science 335, 936–941 (2012).
Litvak, V. et al. A FOXO3–IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490, 421–425 (2012).
Rouse, B.T. & Sehrawat, S. Immunity and immunopathology to viruses: what decides the outcome? Nat. Rev. Immunol. 10, 514–526 (2010).
Navarini, A.A. et al. Increased susceptibility to bacterial superinfection as a consequence of innate antiviral responses. Proc. Natl. Acad. Sci. USA 103, 15535–15539 (2006).
Nakamura, S., Davis, K.M. & Weiser, J.N. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 121, 3657–3665 (2011).
Shahangian, A. et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 119, 1910–1920 (2009).
Foster, S.L., Hargreaves, D.C. & Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007).
Fang, T.C. et al. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J. Exp. Med. 209, 661–669 (2012).
Satoh, T. et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 (2010).
Allan, R.S. et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 487, 249–253 (2012).
Dillon, S.C., Zhang, X., Trievel, R.C. & Cheng, X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6, 227 (2005).
Matsui, T. et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010).
Ceol, C.J. et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517 (2011).
Xu, P.F. et al. Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc. Natl. Acad. Sci. USA 107, 2521–2526 (2010).
Falandry, C. et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J. Biol. Chem. 285, 20234–20241 (2010).
Hogarth, C.A., Mitchell, D., Evanoff, R., Small, C. & Griswold, M. Identification and expression of potential regulators of the mammalian mitotic-to-meiotic transition. Biol. Reprod. 84, 34–42 (2011).
Richon, V.M. et al. Chemogenetic analysis of human protein methyltransferases. Chem. Biol. Drug Des. 78, 199–210 (2011).
Takaoka, A. et al. Cross talk between interferon-γ and -α/β signaling components in caveolar membrane domains. Science 288, 2357–2360 (2000).
Black, J.C., Van Rechem, C. & Whetstine, J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491–507 (2012).
Mantovani, A., Cassatella, M.A., Costantini, C. & Jaillon, S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 11, 519–531 (2011).
McNamee, L.A. & Harmsen, A.G. Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infect. Immun. 74, 6707–6721 (2006).
Cai, S., Batra, S., Lira, S.A., Kolls, J.K. & Jeyaseelan, S. CXCL1 regulates pulmonary host defense to Klebsiella Infection via CXCL2, CXCL5, NF-κB, and MAPKs. J. Immunol. 185, 6214–6225 (2010).
Levy, D. et al. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat. Immunol. 12, 29–36 (2011).
Zhang, Y. et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat. Genet. 34, 181–186 (2003).
Ruthenburg, A.J., Li, H., Patel, D.J. & Allis, C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Marazzi, I. et al. Suppression of the antiviral response by an influenza histone mimic. Nature 483, 428–433 (2012).
Nauseef, W.M. & Borregaard, N. Neutrophils at work. Nat. Immunol. 15, 602–611 (2014).
Raquil, M.A., Anceriz, N., Rouleau, P. & Tessier, P.A. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J. Immunol. 180, 3366–3374 (2008).
Arredouani, M. et al. The scavenger receptor MARCO is required for lung defense against pneumococcal pneumonia and inhaled particles. J. Exp. Med. 200, 267–272 (2004).
Dela Cruz, C.S. et al. Chitinase 3-like-1 promotes Streptococcus pneumoniae killing and augments host tolerance to lung antibacterial responses. Cell Host Microbe 12, 34–46 (2012).
Jamieson, A.M. et al. Role of tissue protection in lethal respiratory viral-bacterial coinfection. Science 340, 1230–1234 (2013).
Helin, K. & Dhanak, D. Chromatin proteins and modifications as drug targets. Nature 502, 480–488 (2013).
Müller, U. et al. Functional role of type I and type II interferons in antiviral defense. Science 264, 1918–1921 (1994).
Honda, K. et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 (2005).
Durbin, J.E., Hackenmiller, R., Simon, M.C. & Levy, D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450 (1996).
Takeuchi, O. et al. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity 11, 443–451 (1999).
Febbraio, M. et al. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J. Clin. Invest. 105, 1049–1056 (2000).
Lee, E.C. et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 (2001).
Sheehan, K.C. et al. Blocking monoclonal antibodies specific for mouse IFN-α/β receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26, 804–819 (2006).
Kratz, P.A., Bottcher, B. & Nassal, M. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc. Natl. Acad. Sci. USA 96, 1915–1920 (1999).
Schebesta, A. et al. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity 27, 49–63 (2007).
Acknowledgements
We thank the Superti-Furga laboratory (Research Center for Molecular Medicine of the Austrian Academy of Sciences) for antibody to Zbp1; S. Schüchner and E. Ogris (Max F. Perutz Laboratories Monoclonal antibody facility in Vienna) for help with generating the antibody to Setdb2; E. Southon and M.E. Palko for technical help in generating the Setdb2GT/GT mice; Y. Guo for technical help; B. Marzolf and P. Troisch (microarray core facility of the Institute for Systems Biology) for technical help in generating microarray data; M. Farlik, T. Penz, M. Schuster and C. Bock (Biosequencing Facility of the Research Center for Molecular Medicine of the Austrian Academy of Sciences) for technical help and advice for generating RNA-Seq data; M. Müller and B. Strobl (University of Veterinary Medicine Vienna) for Irf7−/− and Stat1−/− mice; M. Gorna, F. Grebien, L. Heinz, T. Karonitsch, M. Rebsamen, C. Rosenberger and S. Saluzzo for advice; and D. Barlow, R.A. Flavell, A.N. Hegazy, A. Jamieson, R. Medzhitov and G. Superti-Furga for discussions. Supported by the German Academic Exchange Service (C.S.), the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, of the US National Institutes of Health (E.K.F., A.L.T., L.T. and D.E.S.), the European Molecular Biology Organization (ALTF 48-2008 to A.Be.; and ALTF 314-2012 to R.K.K.), the Austrian Academy of Sciences (A.Bh.), the Swiss National Science Foundation (PP00P3_152928 to D.M.), the Klaus-Tschira Foundation (D.M.), the Gebert-Rüf Foundation (D.M.), the Ohio State University Comprehensive Cancer Center (D.E.S.), the US National Institutes of Health (R01AI032972, R01AI025032 and U19AI100627 to A.A.), the Swiss Foundation for Medical-Biological Stipends (A.Be.), the Austrian Academy of Sciences (A.Be.) and the Austrian Science Fund (P-25360 to A.Be.).
Author information
Authors and Affiliations
Contributions
C.S. designed experiments, performed in vitro and in vivo studies and wrote the manuscript; E.K.F., A.L.T., L.T. and D.E.S. generated the Setdb2GT/GT mouse and provided advice; B.V. performed ChIP, flow cytometry and in vivo experiments; U.R. performed in vitro experiments and generated the monoclonal antibody to Setdb2; S.S., L.B., I.M.G., L.K., A.L., A.Bh. and K.K. performed in vitro experiments; B.B. and M.M.E. performed in vivo experiments and provided advice; R.K.K. performed bioinformatics analyses; F.S., V.L., J.S. and S. Ku. provided reagents and/or advice; I.M., A.H., D.M. and S.Kn. did histological analyses and provided reagents and/or advice; D.E.S. and A.A. supervised the study and provided reagents and advice; and A.Be. supervised the study, designed experiments, performed in vitro and in vivo experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
C.S. and A.Be. are the authors of a patent application related to this work.
Integrated supplementary information
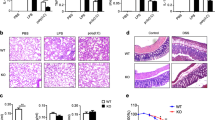
Supplementary Figure 1 Induction of Setdb2 and Zbp1 by PAM3 is TLR2 dependent.
WT and Tlr2–/–Cd36–/– BMDMs were treated with PAM3 (two different batches from Invivogen), LPS or left unstimulated (US). (a) Setdb2 or (b) Zbp1 mRNA expression was quantified by real-time PCR and displayed as fold induction compared to untreated WT cells 8 h after stimulation. The experiment was performed in biological triplicates (mean ± s.e.m.). Statistical significance was calculated by unpaired t-test. Significant p-values were indicated as follows: * p≤0.001, ** p≤0.0001.
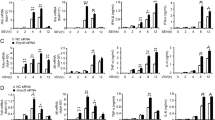
Supplementary Figure 2 Expression kinetics of PKMT-encoding genes in BMDMs after stimulation with poly(I:C).
WT BMDMs were stimulated with poly(I:C) and gene expression levels were determined at the indicated time points by microarray. Fold gene induction compared to unstimulated cells is shown. The top three up-regulated genes are highlighted. The data is derived from systemsimmunology.org (Supplementary Table 3).
Supplementary Figure 3 Generation of Setdb2GT/GT mice.
(a) Schematic of recombinant Setdb2 genetrap targeting strategy. (b) Southern blot of transfected ES cells. (c) Genotyping PCR of Setdb2 genetrap (Setdb2GT/GT) mice.
Supplementary Figure 4 No difference between poly(I:C)-stimulated wild-type and Setdb2GT/GT BMDMs in the degradation of IκBα.
WT and Setdb2GT/GT BMDMs were stimulated with poly(I:C) for indicated times points and IκBα expression/degradation was analyzed by immunoblot using the antibody sc-371. Immunoblot against Actin served as loading control. One out of two similar experiments is shown.
Supplementary Figure 5 Blockade of IFNAR1 leads to increased CXCL1 expression upon stimulation with poly(I:C).
WT and Setdb2GT/GT BMDMs were treated with 20μg/ml IFNAR1-specific antibody (clone MAR1-5A3, anti-IFNAR) or with isotype control followed by poly(I:C) stimulation. (a) Setdb2 and (b) Cxcl1 expression was quantified by real-time PCR after 2 h of stimulation. Relative expression compared to the housekeeping gene Ef1a is shown. The experiment was performed in biological triplicates (mean ± s.e.m.). One out of two similar experiments is shown. Statistical significance was calculated by unpaired t-test. Significant p-values were indicated as follows: * p≤0.01, ** p≤0.001.
Supplementary Figure 6 Time kinetics of the expression of Cxcl1 and Setdb2 in BMDMs after stimulation with poly(I:C).
WT BMDMs were stimulated with poly(I:C) and Cxcl1 and Setdb2 mRNA expression was determined by microarray at the indicated time points. The data was derived from systemsimmunology.org.
Supplementary Figure 7 Setdb2GT/GT alveolar macrophages express increased levels of Cxcl1 and CXCL1 upon infection with influenza virus.
Alveolar macrophages derived from the BAL fluid of naïve WT and Setdb2GT/GT mice were seeded on 96-well tissue culture plates and subsequently infected with influenza virus (PR8) (MOI 100) or left uninfected (UI). 12 h after stimulation, (a) Cxcl1 mRNA expression was quantified by real-time PCR and is displayed as fold induction compared to untreated WT cells. (b) CXCL1 protein was quantified by ELISA. Results from one out of two similar experiments are shown (mean ± s.e.m.). Statistical significance was calculated by unpaired t-test. Significant p-values were indicated as follows: * p≤0.05, ** p≤0.01, *** p≤0.001.
Supplementary Figure 8 Cytokine profiling of mice infected with influenza virus or S. pneumoniae or superinfected with both pathogens.
WT and Setdb2GT/GT mice were either left uninfected (UI), infected with Streptococcus pneumoniae (SP) (~2x104 CFU), infected with a sublethal dose of influenza virus (PR8), or superinfected with SP (~2x104 CFU) (PR8+SP) on day 5 after PR8 infection. BAL fluid was harvested 16 h after SP infection respectively 5 d and 16 h for the groups infected with PR8 or PR8+SP. Levels of (a) Cxcl2, (b) IL-6, (c) IL-10 and (d) CXCL1 were determined by ELISA. Scatter blots represent individual mice (mean ± s.e.m.). (a-c) Results from two pooled experiments are shown. No statistically significant differences between WT and Setdb2GT/GT groups of the respective infections were detected by unpaired t-test.
Supplementary Figure 9 Cellular lung profiling of wild-type and Setdb2GT/GT mice.
WT and Setdb2GT/GT mice were either (a-b) left uninfected (UI), (c-d) infected with influenza virus for 5 d and 16 h, (e-f) infected with SP for 16 h, or (g-h) superinfected with SP for 16 h on day 5 after influenza virus infection (compare Supplementary Fig. 8 legend for respective infectious doses). (a, c, e, g) Representative FACS plots with gating strategies for neutrophils (Neutr), monocytes/macrophages/dendritic cells (Mac/DC), alveolar macrophages (AM), NK cells (NK), T cells and B cells are shown. Scatter plots represent total cell numbers per lung from individual mice. Mean gate frequencies ± s.e.m. are displayed within the FACS plots. (b, d, f, h) Scatter plots represent relative percentages of live CD45+ cells in BAL fluid of individual mice. (i) Representative backgating plot of the population of AMs from CD45+ live lung cells. Scatter blots represent data derived from individual mice (mean ± s.e.m.). (a-d, f-h) Pooled data of two or more experiments are shown. Statistical significance was calculated by unpaired t-test. Significant p-values were indicated as follows: * p≤0.01, ** p≤0.01, *** p≤0.0001.
Supplementary Figure 10 No difference in pathogen loads of Setdb2GT/GT mice and those of wild-type mice after single infection with either influenza virus or S. pneumoniae.
(a) WT and Setdb2GT/GT mice were infected by a sublethal dose of influenza virus (PR8). 5 d later, mice were sacrificed and total RNA was prepared from the right lung lobes. Viral loads were quantified by real-time PCR for the M gene. (b) WT and Setdb2GT/GT mice were infected with ~2x103 respectively ~4x104 CFU of SP. 2 d later, mice were sacrificed and bacterial burden was analyzed by colony formation assay on blood agar plates. Lysates from total lung tissue were analyzed. Scatter blots represent data derived from individual mice pooled from 1-3 independent experiments (mean ±_s.e.m.). No statistically significant differences between WT and Setdb2GT/GT groups of the respective infections were detected by unpaired t-test.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–3 (PDF 1431 kb)
Supplementary Table 1
Microarray data of lung tissue from influenza virus infected compared to mock treated wild type mice. (XLSX 94 kb)
Supplementary Table 2
Predicted transcription factor binding sites of Setdb2. (XLSX 65 kb)
Supplementary Table 3
Microarray data of poly(I:C) stimulated wild type BMDMs (XLSX 48 kb)
Supplementary Table 4
RNAseq data of poly(I:C) stimulated WT and Setdb2GT/GT BMDMs. (XLSX 95 kb)
Supplementary Table 5
List of NF-κB target genes (XLSX 37 kb)
Rights and permissions
About this article
Cite this article
Schliehe, C., Flynn, E., Vilagos, B. et al. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat Immunol 16, 67–74 (2015). https://doi.org/10.1038/ni.3046
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3046
This article is cited by
-
RETRACTED ARTICLE: Selected TLR7/8 agonist and type I interferon (IFN-α) cooperatively redefine the microglia transcriptome
Inflammopharmacology (2023)
-
Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance
Nature Reviews Molecular Cell Biology (2022)
-
Dendritic cell migration in inflammation and immunity
Cellular & Molecular Immunology (2021)
-
Innate immune responses in RNA viral infection
Frontiers of Medicine (2021)
-
Methyltransferase Dot1l preferentially promotes innate IL-6 and IFN-β production by mediating H3K79me2/3 methylation in macrophages
Cellular & Molecular Immunology (2020)