Abstract
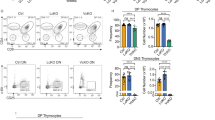
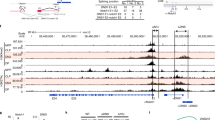
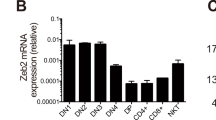
E proteins are basic helix-loop-helix transcription factors that regulate many key aspects of lymphocyte development. Thymocytes express multiple E proteins that are thought to provide cooperative and compensatory functions crucial for T cell differentiation. Contrary to that, we report here that the E protein HEB was uniquely required at the CD4+CD8+ double-positive (DP) stage of T cell development. Thymocytes lacking HEB showed impaired survival, failed to make rearrangements of variable-α (Vα) segments to distal joining-α (Jα) segments in the gene encoding the T cell antigen receptor α-chain (Tcra) and had a profound, intrinsic block in the development of invariant natural killer T cells (iNKT cells) at their earliest progenitor stage. Thus, our results show that HEB is a specific and essential factor in T cell development and in the generation of the iNKT cell lineage, defining a unique role for HEB in the regulation of lymphocyte maturation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
12 March 2010
In the version of this article initially published, references 35–41 were cited out of order in the text. The citations on page 247 should be as follows: column one, first paragraph, second full sentence, “RAG-2 recombinase protein35,36, which could in turn impair rearrangements37”; column two, middle paragraph, final two sentences, “iNKT development20-23,28,29,38,39....from stage 0 through stage 2 (refs. 20,38)”; column two, final paragraph, second sentence, “RORγt-deficient thymocytes40”; and column two, final paragraph, fourth sentence, “Tcra rearrangements33,34,41.” The reference list should be as follows: 37. Yannoutsos, N. et al. The role of recombination activating gene (RAG) reinduction in thymocyte development in vivo. J. Exp. Med. 194, 471–480 (2001). 38. Nichols, K.E. et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 11, 340–345 (2005). 39. Savage, A.K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008). 40. Benlagha, K., Wei, D.G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005). 41. Sun, Z. et al. Requirement for RORγ in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 (2000). The error has been corrected in the HTML and PDF versions of the article.
References
Engel, I., Johns, C., Bain, G., Rivera, R.R. & Murre, C. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194, 733–745 (2001).
Murre, C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6, 1079–1086 (2005).
Cisse, B. et al. Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48 (2008).
Ikawa, T., Kawamoto, H., Goldrath, A.W. & Murre, C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 203, 1329–1342 (2006).
Rolf, J. et al. Molecular profiling reveals distinct functional attributes of CD1d-restricted natural killer (NK) T cell subsets. Mol. Immunol. 45, 2607–2620 (2008).
Engel, I. & Murre, C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 23, 202–211 (2004).
Jones, M.E. & Zhuang, Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 27, 860–870 (2007).
Barndt, R., Dai, M.F. & Zhuang, Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during αβ thymopoiesis. J. Immunol. 163, 3331–3343 (1999).
Barndt, R.J., Dai, M. & Zhuang, Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 20, 6677–6685 (2000).
Bain, G. et al. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 17, 4782–4791 (1997).
Wojciechowski, J., Lai, A., Kondo, M. & Zhuang, Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J. Immunol. 178, 5717–5726 (2007).
Zhuang, Y., Barndt, R.J., Pan, L., Kelley, R. & Dai, M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol. Cell. Biol. 18, 3340–3349 (1998).
Ravanpay, A.C. & Olson, J.M. E protein dosage influences brain development more than family member identity. J. Neurosci. Res. 86, 1472–1481 (2008).
Godfrey, D.I. & Berzins, S.P. Control points in NKT-cell development. Nat. Rev. Immunol. 7, 505–518 (2007).
Geissmann, F. et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 3, e113 (2005).
Matsuda, J.L., Mallevaey, T., Scott-Browne, J. & Gapin, L. CD1d-restricted iNKT cells, the ′Swiss-Army knife′ of the immune system. Curr. Opin. Immunol. 20, 358–368 (2008).
Tupin, E., Kinjo, Y. & Kronenberg, M. The unique role of natural killer T cells in the response to microorganisms. Nat. Rev. Microbiol. 5, 405–417 (2007).
Bendelac, A., Savage, P.B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Kronenberg, M. & Engel, I. On the road: progress in finding the unique pathway of invariant NKT cell differentiation. Curr. Opin. Immunol. 19, 186–193 (2007).
Dose, M. et al. Intrathymic proliferation wave essential for Vα14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA 106, 8641–8646 (2009).
Mycko, M.P. et al. Selective requirement for c-Myc at an early stage of Vα14i NKT cell development. J. Immunol. 182, 4641–4648 (2009).
Bezbradica, J.S., Hill, T., Stanic, A.K., Van Kaer, L. & Joyce, S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc. Natl. Acad. Sci. USA 102, 5114–5119 (2005).
Egawa, T. et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Hager, E., Hawwari, A., Matsuda, J.L., Krangel, M.S. & Gapin, L. Multiple constraints at the level of TCRα rearrangement impact Vα14i NKT cell development. J. Immunol. 179, 2228–2234 (2007).
Matsuda, J.L. et al. T-bet concomitantly controls migration, survival, and effector functions during the development of Vα14i NKT cells. Blood 107, 2797–2805 (2006).
Townsend, M.J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity 20, 477–494 (2004).
Ohteki, T., Ho, S., Suzuki, H., Mak, T.W. & Ohashi, P.S. Role for IL-15/IL-15 receptor β-chain in natural killer 1.1+ T cell receptor-αβ+ cell development. J. Immunol. 159, 5931–5935 (1997).
Lazarevic, V. et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10, 306–313 (2009).
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 (2008).
Mattner, J. et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe 3, 304–315 (2008).
Pan, L., Hanrahan, J., Li, J., Hale, L.P. & Zhuang, Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J. Immunol. 168, 3923–3932 (2002).
Wolfer, A. et al. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2, 235–241 (2001).
Xi, H., Schwartz, R., Engel, I., Murre, C. & Kersh, G.J. Interplay between RORγt, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity 24, 813–826 (2006).
Guo, J. et al. Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat. Immunol. 3, 469–476 (2002).
Li, Z., Dordai, D.I., Lee, J. & Desiderio, S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity 5, 575–589 (1996).
Lin, W.C. & Desiderio, S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc. Natl. Acad. Sci. USA 91, 2733–2737 (1994).
Yannoutsos, N. et al. The role of recombination activating gene (RAG) reinduction in thymocyte development in vivo. J. Exp. Med. 194, 471–480 (2001).
Nichols, K.E. et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 11, 340–345 (2005).
Savage, A.K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008).
Benlagha, K., Wei, D.G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005).
Sun, Z. et al. Requirement for RORγ in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 (2000).
Bendelac, A., Hunziker, R.D. & Lantz, O. Increased interleukin 4 and immunoglobuline E production in transgenic mice overexpressing NK1 T cells. J. Exp. Med. 184, 1285–1293 (1996).
Tupin, E. & Kronenberg, M. Activation of natural killer T cells by glycolipids. Methods Enzymol. 417, 185–201 (2006).
Riegert, P. & Gilfillan, S. A conserved sequence block in the murine and human TCR Jα region: assessment of regulatory function in vivo. J. Immunol. 162, 3471–3480 (1999).
Pasqual, N. et al. Quantitative and qualitative changes in V-Jα rearrangements during mouse thymocytes differentiation: implication for a limited T cell receptor α chain repertoire. J. Exp. Med. 196, 1163–1173 (2002).
Acknowledgements
We thank C. Murre (University of California San Diego) for discussions, reagents and mice; Y. Zhuang (Duke University) for Tcf12−/−, Tcf12f/f and Tcfe2af/f mouse lines; A. Bendelac (Howard Hughes Medical Institute) and M. Kronenberg (La Jolla Institute for Allergy and Immunology) for Vα14-Jα18 TCR–transgenic mice; A. Abbas (University of California San Francisco) for the constructs MSCV2.2–Bcl-xL–IRES–Thy-1.1 and MSCV2.2–empty–IRES–Thy-1.1; M. Kronenberg for advice; I. Ch'en for advice and assistance; and J. Hamerman, G. Barton, E. Zuniga and S. Hedrick and members of the Goldrath laboratory for critical review of the manuscript. Supported by the Cancer Research Institute, the Pew Charitable Trusts, the National Institutes of Health (AI067545 and AI072117 to A.W.G.), the Leukemia & Lymphoma Society (L.M.D.) and University of California San Diego undergraduate research scholarship program (J.K.F.).
Author information
Authors and Affiliations
Contributions
L.M.D. designed the study, did experiments, analyzed data and wrote the manuscript; J.K. and J.K.F. did experiments and analyzed data; and A.W.G. designed the study, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1–2 (PDF 3041 kb)
Rights and permissions
About this article
Cite this article
D'Cruz, L., Knell, J., Fujimoto, J. et al. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol 11, 240–249 (2010). https://doi.org/10.1038/ni.1845
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1845
This article is cited by
-
microRNA dynamic expression regulates invariant NKT cells
Cellular and Molecular Life Sciences (2021)
-
Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge
Nature Reviews Immunology (2020)
-
Transcription factor YY1 is essential for iNKT cell development
Cellular & Molecular Immunology (2019)
-
TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4+CD8+ thymocytes
Nature Immunology (2018)
-
Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70
Nature Communications (2018)