Abstract
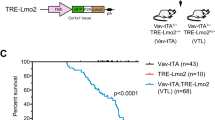
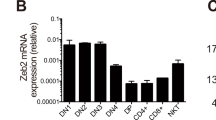
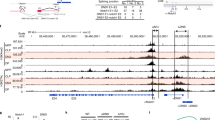
T lymphocyte acute lymphoblastic leukemia (T-ALL) is frequently associated with increased expression of the E protein transcription factor inhibitors TAL1 and LYL1. In mouse models, ectopic expression of TAL1 or LYL1 in T cell progenitors, or inactivation of E2A, is sufficient to predispose mice to develop T-ALL. How E2A suppresses thymocyte transformation is currently unknown. Here, we show that early deletion of E2a, prior to the DN3 stage, was required for robust leukemogenesis and was associated with alterations in thymus cellularity, T cell differentiation, and gene expression in immature CD4+CD8+ thymocytes. Introduction of wild-type thymocytes into mice with early deletion of E2a prevented leukemogenesis, or delayed disease onset, and impacted the expression of multiple genes associated with transformation and genome instability. Our data indicate that E2A suppresses leukemogenesis by promoting T cell development and enforcing inter-thymocyte competition, a mechanism that is emerging as a safeguard against thymocyte transformation. These studies have implications for understanding how multiple essential regulators of T cell development suppress T-ALL and support the hypothesis that thymocyte competition suppresses leukemogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The RNA-seq data presented in this paper are available through the Gene Expression Omnibus database under accession number GSE234609 and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE234609. Other data in the paper will be made available upon reasonable request.
References
Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–90.
Noronha EP, Marques LVC, Andrade FG, Thuler LCS, Terra-Granado E, Pombo-de-Oliveira MS, et al. The Profile of Immunophenotype and Genotype Aberrations in Subsets of Pediatric T-Cell Acute Lymphoblastic Leukemia. Front Oncol. 2019;9:316.
Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16:494–507.
Ferrando AA, Look AT. Gene expression profiling in T-cell acute lymphoblastic leukemia. Semin Hematol. 2003;40:274–80.
Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–56.
O’Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–5.
Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Investig. 2012;122:3398–406.
Parriott G, Kee BL. E Protein Transcription Factors as Suppressors of T Lymphocyte Acute Lymphoblastic Leukemia. Front Immunol. 2022;13:885144.
Bain G, Engel I, Robanus Maandag EC, te Riele HPJ, Voland JR, Sharp LL, et al. E2A Deficiency Leads to Abnormalities in αβ T-Cell Development and to Rapid Development of T-Cell Lymphomas. Mol Cell Biol. 1997;17:4782–91.
Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High Incidence of T-Cell Tumors in E2A-Null Mice and E2A/Id1 Double-Knockout Mice. Mol Cell Biol. 1997;17:7317–27.
Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, et al. Combinatorial Transcriptional Control In Blood Stem/Progenitor Cells: Genome-wide Analysis of Ten Major Transcriptional Regulators. Cell Stem Cell. 2010;7:532–44.
de Pooter RF, Dias S, Chowdhury M, Bartom ET, Okoreeh MK, Sigvardsson M, et al. Cutting Edge: Lymphomyeloid-Primed Progenitor Cell Fates Are Controlled by the Transcription Factor Tal1. J Immunol. 2019;202:2837–42.
Zohren F, Souroullas GP, Luo M, Gerdemann U, Imperato MR, Wilson NK, et al. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol. 2012;13:761–9.
Bouderlique T, Peña-Pérez L, Kharazi S, Hils M, Li X, Krstic A, et al. The Concerted Action of E2-2 and HEB Is Critical for Early Lymphoid Specification. Front Immunol. 2019;10:455.
Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014;14:529–45.
Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol Rev. 2006;209:191–211.
Tan TK, Zhang C, Sanda T. Oncogenic transcriptional program driven by TAL1 in T-cell acute lymphoblastic leukemia. Int J Hematol. 2019;109:5–17.
O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004;5:587–96.
Kim D, Peng XC, Sun XH. Massive Apoptosis of Thymocytes in T-Cell-Deficient Id1 Transgenic Mice. Mol Cell Biol. 1999;19:8240–53.
Morrow MA, Mayer EW, Perez CA, Adlam M, Siu G. Overexpression of the Helix–Loop–Helix protein Id2 blocks T cell development at multiple stages. Mol Immunol. 1999;36:491–503.
Paiva RA, Ramos CV, Martins VC. Thymus autonomy as a prelude to leukemia. FEBS J. 2018;285:4565–74.
Peaudecerf L, Lemos S, Galgano A, Krenn G, Vasseur F, Di Santo JP, et al. Thymocytes may persist and differentiate without any input from bone marrow progenitors. J Exp Med. 2012;209:1401–8.
Martins VC, Ruggiero E, Schlenner SM, Madan V, Schmidt M, Fink PJ, et al. Thymus-autonomous T cell development in the absence of progenitor import. J Exp Med. 2012;209:1409–17.
Martins VC, Busch K, Juraeva D, Blum C, Ludwig C, Rasche V, et al. Cell competition is a tumour suppressor mechanism in the thymus. Nature. 2014;509:465–70.
Abdulla HD, Alserihi R, Flensburg C, Abeysekera W, Luo MX, Gray DHD, et al. Overexpression of Lmo2 initiates T-lymphoblastic leukemia via impaired thymocyte competition. J Exp Med. 2023;220:e20212383.
McCormack MP, Shields BJ, Jackson JT, Nasa C, Shi W, Slater NJ, et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood. 2013;122:2093–103.
Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An Analysis of T Cell Intrinsic Roles of E2A by Conditional Gene Disruption in the Thymus. J Immunol. 2002;168:3923–32.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Manning J, WackerO, Peltzer A, Nf-Core Bot, Ribeiro-Dantas M, Harshil Patel, et al. nf-core/differentialabundance: v1.2.0 - 2023-04-19. Zenodo; 2023 [cited 2023 Apr 22]. https://zenodo.org/record/7568000.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Carr T, McGregor S, Dias S, Verykokakis M, Le Beau MM, Xue HH, et al. Oncogenic and Tumor Suppressor Functions for Lymphoid Enhancer Factor 1 in E2a-/- T Acute Lymphoblastic Leukemia. Front Immunol. 2022;13:845488.
Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, et al. Notch Signaling Augments T Cell Responsiveness by Enhancing CD25 Expression. J Immunol. 2003;171:2896–903.
Dias S, Månsson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity. 2008;29:217–27.
Krishnamoorthy V, Carr T, de Pooter RF, Akinola EO, Gounari F, Kee BL. Repression of Ccr9 Transcription in Mouse T Lymphocyte Progenitors by the Notch Signaling Pathway. J Immunol. 2015;194:3191–200.
Yang Q, Esplin B, Borghesi L. E47 regulates hematopoietic stem cell proliferation and energetics but not myeloid lineage restriction. Blood. 2011;117:3529–38.
Kee BL, Bain G, Murre C. IL-7Ralpha and E47: independent pathways required for development of multipotent lymphoid progenitors. EMBO J. 2002;21:103–13.
Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–42.
Xu W, Carr T, Ramirez K, McGregor S, Sigvardsson M, Kee BL. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood. 2013;121:1534–42.
Qian L, Bajana S, Georgescu C, Peng V, Wang HC, Adrianto I, et al. Suppression of ILC2 differentiation from committed T cell precursors by E protein transcription factors. J Exp Med. 2019;216:884–99.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109.
Reschly EJ, Spaulding C, Vilimas T, Graham WV, Brumbaugh RL, Aifantis I, et al. Notch1 promotes survival of E2A-deficient T cell lymphomas through pre–T cell receptor–dependent and –independent mechanisms. Blood. 2006;107:4115–21.
Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCFFbw7 complex in development and transformation. Oncogene. 2010;29:4865–73.
Laptenko O, Prives C. p53: master of life, death, and the epigenome. Genes Dev. 2017;31:955–6.
Hurlin PJ, Quéva C, Koskinen PJ, Steingrímsson E, Ayer DE, Copeland NG, et al. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1996;15:2030.
Hu CL, Chen BY, Li Z, Yang T, Xu CH, Yang R, et al. Targeting UHRF1-SAP30-MXD4 axis for leukemia initiating cell eradication in myeloid leukemia. Cell Res. 2022;32:1105–23.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating Mutations of NOTCH1 in Human T Cell Acute Lymphoblastic Leukemia. Science. 2004;306:269–71.
Elias HK, Bryder D, Park CY. Molecular mechanisms underlying lineage bias in aging hematopoiesis. Semin Hematol. 2017;54:4–11.
Bensberg M, Rundquist O, Selimović A, Lagerwall C, Benson M, Gustafsson M, et al. TET2 as a tumor suppressor and therapeutic target in T-cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2021;118:e2110758118.
Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–81.
Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras–initiated leukemia. J Clin Invest. 2008;118:3181–94.
Kashiwagi M, Figueroa DS, Ay F, Morgan BA, Georgopoulos K. A double-negative thymocyte-specific enhancer augments Notch1 signaling to direct early T cell progenitor expansion, lineage restriction and β-selection. Nat Immunol. 2022;23:1628–43.
O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–24.
Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–35.
Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–25.
Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–6.
Chiang MY, Wang Q, Gormley AC, Stein SJ, Xu L, Shestova O, et al. High selective pressure for Notch1 mutations that induce Myc in T-cell acute lymphoblastic leukemia. Blood. 2016;128:2229–40.
Acknowledgements
We thank members of the Kee laboratory, S. Dias, and A. Melnick for helpful discussions and S. Liang for technical assistance. We thank the University of Chicago Genomics Facility (RRID: SCR_019196) and Cytometry and Antibody Technology Facility (RRID: SCR_017760).
Funding
This work was funded by the National Institutes of Health grants R21 AI119894, R01 AI107213, the Janet Rowley Fund from the University of Chicago Comprehensive Cancer Center, and a Team Science award from the Biological Science Division of the University of Chicago to BLK. GP was supported by T32 AI007090. The University of Chicago Genomics Facility and Cytometry and Antibody Technology Facility receive financial support from the Cancer Center Support Grant (P30 CA014599), and the Animal Resource Center.
Author information
Authors and Affiliations
Contributions
GP designed, performed, and interpreted experiments and analyzed RNA-seq data, EH, REM and CF maintained mice, performed and analyzed experiments, and had essential intellectual input, CS and WS provided valuable advice and edited the manuscript, ETB performed RNA-seq analysis and edited the manuscript, BLK conceived of the project, designed and interpreted experiments, and provided funding for the project. GP and BLK wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Parriott, G., Hegermiller, E., Morman, R.E. et al. Loss of thymocyte competition underlies the tumor suppressive functions of the E2a transcription factor in T-ALL. Leukemia 38, 491–501 (2024). https://doi.org/10.1038/s41375-023-02123-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-02123-4