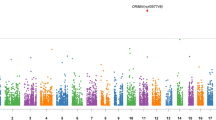
Abstract
Despite a substantial genetic component, efforts to identify common genetic variation underlying depression have largely been unsuccessful. In the current study we aimed to identify rare genetic variants that might have large effects on depression in the general population. Using high-coverage exome-sequencing, we studied the exonic variants in 1265 individuals from the Rotterdam study (RS), who were assessed for depressive symptoms. We identified a missense Asn396Ser mutation (rs77960347) in the endothelial lipase (LIPG) gene, occurring with an allele frequency of 1% in the general population, which was significantly associated with depressive symptoms (P-value=5.2 × 10−08, β=7.2). Replication in three independent data sets (N=3612) confirmed the association of Asn396Ser (P-value=7.1 × 10−03, β=2.55) with depressive symptoms. LIPG is predicted to have enzymatic function in steroid biosynthesis, cholesterol biosynthesis and thyroid hormone metabolic processes. The Asn396Ser variant is predicted to have a damaging effect on the function of LIPG. Within the discovery population, carriers also showed an increased burden of white matter lesions (P-value=3.3 × 1−02) and a higher risk of Alzheimer’s disease (odds ration=2.01; P-value=2.8 × 10−02) compared with the non-carriers. Together, these findings implicate the Asn396Ser variant of LIPG in the pathogenesis of depressive symptoms in the general population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lohoff FW . Overview of the genetics of major depressive disorder. Curr Psychiatry Rep 2010; 12: 539–546.
Visscher PM, Brown MA, McCarthy MI, Yang J . Five years of GWAS discovery. Am J Hum Genet 2012; 90: 7–24.
Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013; 18: 497–511.
CONVERGE Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 2015; 523: 588–591.
Sullivan PF . Genetics of disease: associations with depression. Nature 2015; 523: 539–540.
Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR et al. Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol Psychiatry 2014; 76: 510–512.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427.
Demirkan A, Lahti J, Direk N, Viktorin A, Lunetta KL, Terracciano A et al. Somatic, positive and negative domains of the CES-D scale: a meta-analysis of genome-wide association studies. Psychol Med 2015; 46: 1613–1623.
Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC et al. A genome-wide association study of depressive symptoms. Biol Psychiatry 2013; 73: 667–678.
Gottesman II, Gould TD . The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 2003; 160: 636–645.
Hall MH, Smoller JW . A new role for endophenotypes in the GWAS era: functional characterization of risk variants. Harv Rev Psychiatry 2010; 18: 67–74.
International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011; 478: 103–109.
Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994.
Ott J, Wang J, Leal SM . Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet 2015; 16: 275–284.
Holmans P, Zubenko GS, Crowe RR, DePaulo JR Jr., Scheftner WA, Weissman MM et al. Genomewide significant linkage to recurrent, early-onset major depressive disorder on chromosome 15q. Am J Hum Genet 2004; 74: 1154–1167.
Levinson DF, Evgrafov OV, Knowles JA, Potash JB, Weissman MM, Scheftner WA et al. Genetics of recurrent early-onset major depression (GenRED): significant linkage on chromosome 15q25-q26 after fine mapping with single nucleotide polymorphism markers. Am J Psychiatry 2007; 164: 259–264.
Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, Depaulo JR Jr. et al. Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am J Psychiatry 2007; 164: 248–258.
Camp NJ, Lowry MR, Richards RL, Plenk AM, Carter C, Hensel CH et al. Genome-wide linkage analyses of extended Utah pedigrees identifies loci that influence recurrent, early-onset major depression and anxiety disorders. Am J Med Genet B Neuropsychiatr Genet 2005; 135B: 85–93.
Abkevich V, Camp NJ, Hensel CH, Neff CD, Russell DL, Hughes DC et al. Predisposition locus for major depression at chromosome 12q22-12q23.2. Am J Hum Genet 2003; 73: 1271–1281.
McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N et al. Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet 2005; 14: 3337–3345.
McCarthy S, Das S, Kretzschmar W, Durbin R, Abecasis G, Marchini J . A reference panel of 64,976 haplotypes for genotype imputation. bioRxiv 2015; http://dx.doi.org/10.1101/035170.
Radloff LS . The CES-D scale: a self report depression scale for research in the general population. Appl Pshycol Measurement 1977; 1: 385–401.
Hofman A, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, Ikram MA et al. The Rotterdam Study: 2014 objectives and design update. Eur J Epidemiol 2013; 28: 889–926.
Harlow SD, Goldberg EL, Comstock GW . A longitudinal study of the prevalence of depressive symptomatology in elderly widowed and married women. Arch Gen Psychiatry 1991; 48: 1065–1068.
Kuchibhatla MN, Fillenbaum GG, Hybels CF, Blazer DG . Trajectory classes of depressive symptoms in a community sample of older adults. Acta Psychiatr Scand 2012; 125: 492–501.
Lopez-Leon S, Choy WC, Aulchenko YS, Claes SJ, Oostra BA, Mackenbach JP et al. Genetic factors influence the clustering of depression among individuals with lower socioeconomic status. PLoS ONE 2009; 4: e5069.
Lyness JM, Noel TK, Cox C, King DA, Conwell Y, Caine ED . Screening for depression in elderly primary care patients. A comparison of the Center for Epidemiologic Studies-Depression Scale and the Geriatric Depression Scale. Arch Intern Med 1997; 157: 449–454.
Beekman AT, Deeg DJ, Van Limbeek J, Braam AW, De Vries MZ, Van Tilburg W . Criterion validity of the Center for Epidemiologic Studies Depression scale (CES-D): results from a community-based sample of older subjects in The Netherlands. Psychol Med 1997; 27: 231–235.
Radloff LS . The CED-D scale: a self-report depression scale for research in the general population. Appl Psychol Meas 1977; 1: 385–401.
de Boer R, Vrooman HA, van der Lijn F, Vernooij MW, Ikram MA, van der Lugt A et al. White matter lesion extension to automatic brain tissue segmentation on MRI. Neuroimage 2009; 45: 1151–1161.
Vrooman HA, Cocosco CA, van der Lijn F, Stokking R, Ikram MA, Vernooij MW et al. Multi-spectral brain tissue segmentation using automatically trained k-Nearest-Neighbor classification. Neuroimage 2007; 37: 71–81.
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–1458.
Chaker L, Heeringa J, Dehghan A, Medici M, Visser WE, Baumgartner C et al. Normal thyroid function and the risk of atrial fibrillation: the Rotterdam Study. J Clin Endocrinol Metab 2015; 100: 3718–3724.
Pardo LM, MacKay I, Oostra B, van Duijn CM, Aulchenko YS . The effect of genetic drift in a young genetically isolated population. Ann Hum Genet 2005; 69 (Pt 3): 288–295.
Bjelland I, Dahl AA, Haug TT, Neckelmann D . The validity of the Hospital Anxiety and Depression Scale. An updated literature review. J Psychosom Res 2002; 52: 69–77.
Draisma HH, Pool R, Kobl M, Jansen R, Petersen AK, Vaarhorst AA et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat Commun 2015; 6: 7208.
Gonzalez-Covarrubias V, Beekman M, Uh HW, Dane A, Troost J, Paliukhovich I et al. Lipidomics of familial longevity. Aging Cell 2013; 12: 426–434.
Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet 2012; 8: e1002490.
Demirkan A, Henneman P, Verhoeven A, Dharuri H, Amin N, van Klinken JB et al. Insight in genome-wide association of metabolite quantitative traits by exome sequence analyses. PLoS Genet 2015; 11: e1004835.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. The variant call format and VCFtools. Bioinformatics 2011; 27: 2156–2158.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Brouwer RW, van den Hout MC, Grosveld FG, van Ijcken WF . NARWHAL, a primary analysis pipeline for NGS data. Bioinformatics 2012; 28: 284–285.
Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS ONE 2013; 8: e68095.
Yasuda T, Ishida T, Rader DJ . Update on the role of endothelial lipase in high-density lipoprotein metabolism, reverse cholesterol transport, and atherosclerosis. Circ J 2010; 74: 2263–2270.
Kluz J, Adamiec R . [The role of endothelial lipase in the pathogenesis of atherosclerosis] Rola lipazy srodblonkowej w patogenezie miazdycy. Pol Arch Med Wewn 2006; 115: 148–156.
Huang J, Qian HY, Li ZZ, Zhang JM, Wang S, Tao Y et al. Role of endothelial lipase in atherosclerosis. Transl Res 2010; 156: 1–6.
Paradis ME, Lamarche B . Endothelial lipase: its role in cardiovascular disease. Can J Cardiol 2006; 22 (Suppl B): 31B–34B.
Broedl UC, Jin W, Rader DJ . Endothelial lipase: a modulator of lipoprotein metabolism upregulated by inflammation. Trends Cardiovasc Med 2004; 14: 202–206.
Global Lipids Genetics Consortium, Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013; 45: 1274–1283.
van Reedt Dortland AK, Giltay EJ, van Veen T, van Pelt J, Zitman FG, Penninx BW . Associations between serum lipids and major depressive disorder: results from the Netherlands Study of Depression and Anxiety (NESDA). J Clin Psychiatry 2010; 71: 729–736.
O'Brien JT, Ames D . White matter lesions in depression and Alzheimer’s disease. Br J Psychiatry 1996; 169: 671.
Brown FW, Lewine RJ, Hudgins PA, Risch SC . White matter hyperintensity signals in psychiatric and nonpsychiatric subjects. Am J Psychiatry 1992; 149: 620–625.
Coffey CE, Wilkinson WE, Weiner RD, Parashos IA, Djang WT, Webb MC et al. Quantitative cerebral anatomy in depression. A controlled magnetic resonance imaging study. Arch Gen Psychiatry 1993; 50: 7–16.
de Groot JC, de Leeuw FE, Oudkerk M, Hofman A, Jolles J, Breteler MM . Cerebral white matter lesions and depressive symptoms in elderly adults. Arch Gen Psychiatry 2000; 57: 1071–1076.
Prins ND, van Dijk EJ, den Heijer T, Vermeer SE, Koudstaal PJ, Oudkerk M et al. Cerebral white matter lesions and the risk of dementia. Arch Neurol 2004; 61: 1531–1534.
Fazekas F, Chawluk JB, Alavi A, Hurtig HI, Zimmerman RA . MR signal abnormalities at 1.5T in Alzheimer’s dementia and normal aging. AJR Am J Roentgenol 1987; 149: 351–356.
Yoshita M, Fletcher E, Harvey D, Ortega M, Martinez O, Mungas DM et al. Extent and distribution of white matter hyperintensities in normal aging, MCI, and AD. Neurology 2006; 67: 2192–2198.
Tsuno N, Homma A . What is the association between depression and Alzheimer’s disease? Expert Rev Neurother 2009; 9: 1667–1676.
Andersen K, Lolk A, Kragh-Sorensen P, Petersen NE, Green A . Depression and the risk of Alzheimer disease. Epidemiology 2005; 16: 233–238.
Dal Forno G, Palermo MT, Donohue JE, Karagiozis H, Zonderman AB, Kawas CH . Depressive symptoms, sex, and risk for Alzheimer’s disease. Ann Neurol 2005; 57: 381–387.
Fehrmann RS, Karjalainen JM, Krajewska M, Westra HJ, Maloney D, Simeonov A et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet 2015; 47: 115–125.
Bahls SC, de Carvalho GA . [The relation between thyroid function and depression: a review] A relacao entre a funcao tireoidiana e a depressao: uma revisao. Rev Bras Psiquiatr 2004; 26: 41–49.
Bauer M, Goetz T, Glenn T, Whybrow PC . The thyroid-brain interaction in thyroid disorders and mood disorders. J Neuroendocrinol 2008; 20: 1101–1114.
Joffe RT, Sokolov ST . Thyroid hormone treatment of primary unipolar depression: a review. Int J Neuropsychopharmacol 2000; 3: 143–147.
Medici M, Direk N, Visser WE, Korevaar TI, Hofman A, Visser TJ et al. Thyroid function within the normal range and the risk of depression: a population-based cohort study. J Clin Endocrinol Metab 2014; 99: 1213–1219.
Ogawa S, Fujii T, Koga N, Hori H, Teraishi T, Hattori K et al. Plasma L-tryptophan concentration in major depressive disorder: new data and meta-analysis. J Clin Psychiatry 2014; 75: e906–e915.
Lindseth G, Helland B, Caspers J . The effects of dietary tryptophan on affective disorders. Arch Psychiatr Nurs 2015; 29: 102–107.
Resende WR, Valvassori SS, Reus GZ, Varela RB, Arent CO, Ribeiro KF et al. Effects of sodium butyrate in animal models of mania and depression: implications as a new mood stabilizer. Behav Pharmacol 2013; 24: 569–579.
Harlow SD, Goldberg EL, Comstock GW . A longitudinal study of risk factors for depressive symptomatology in elderly widowed and married women. Am J Epidemiol 1991; 134: 526–538.
van der Sluis S, Posthuma D, Nivard MG, Verhage M, Dolan CV . Power in GWAS: lifting the curse of the clinical cut-off. Mol Psychiatry 2013; 18: 2–3.
Acknowledgements
Erasmus Rucphen family study: The ERF study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the program ‘Quality of Life and Management of the Living Resources’ of the 5th Framework Programme (number QLG2-CT-2002-01254). High-throughput analysis of the ERF data was supported by a joint grant from the Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). Exome sequencing analysis in ERF was supported by the ZonMw grant (project 91111025). Metabolomics was partly funded by BBMRI-NL. We are grateful to all study participants and their relatives, general practitioners and neurologists for their contributions and to P Veraart for her help in genealogy, J Vergeer for the supervision of the laboratory work and P Snijders for his help in data collection. NA is supported by the Netherlands Brain Foundation (project number F2013(1)-28). FMSdV was supported by the Netherlands Brain Foundation (grant number FS2012(1)-39). Rotterdam study: The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. We are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. The generation and management of the exome sequencing data for the Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, The Netherlands. The Exome Sequencing data set was funded by the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) sponsored Netherlands Consortium for Healthy Aging (NCHA; project number 050-060-810), by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC and by the and by a Complementation Project of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL; www.bbmri.nl; project number CP2010-41). Metabolomics assessment was funded by the Rainbow Project 4 of BBMRI-NL. We thank Mr Pascal Arp, Ms Mila Jhamai and Mr Marijn Verkerk for their help in creating the RS-Exome Sequencing database.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Rights and permissions
About this article
Cite this article
Amin, N., Jovanova, O., Adams, H. et al. Exome-sequencing in a large population-based study reveals a rare Asn396Ser variant in the LIPG gene associated with depressive symptoms. Mol Psychiatry 22, 537–543 (2017). https://doi.org/10.1038/mp.2016.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2016.101
This article is cited by
-
Integration of whole-exome sequencing and structural neuroimaging analysis in major depressive disorder: a joint study
Translational Psychiatry (2024)
-
Common and rare variant associations with latent traits underlying depression, bipolar disorder, and schizophrenia
Translational Psychiatry (2023)
-
Sex differences in the genetic architecture of depression
Scientific Reports (2020)
-
Missing heritability in Parkinson’s disease: the emerging role of non-coding genetic variation
Journal of Neural Transmission (2020)
-
Association of plasma endothelial lipase levels on cognitive impairment
BMC Psychiatry (2019)