Abstract
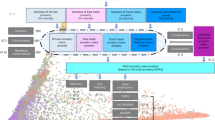
Despite thousands of common genetic loci of major depression disorders (MDD) have been identified by GWAS to date, a large proportion of genetic variation predisposing to MDD remains unaccounted for. By utilizing the newly released UK Biobank 200,643 exome dataset, we conducted an exome-wide association study to identify rare risk variants contributing to MDD. After quality control, 120,033 participants with MDD polygenic risk scores (PRS) values were included. The individuals with lower 30% quantile of the PRS value were filtered for case and control selecting. Then the cases were set as the individuals with upper 10% quantile of the PHQ depression score and lower 10% quantile were set as controls. Finally, 1612 cases and 1612 controls were included in this study. The variants were annotated by ANNOVRA software. After exclusions, 34,761 qualifying variants, including 148 frameshift variant, 335 non-frameshift variant, 33,758 nonsynonymous, 91 start-loss, 393 stop-gain, 36 stop-loss variants were imported into the SKAT R-package to perform single variants, gene-based burden and robust burden tests with minor allele frequency (MAF) < 0.01. Single variant association testing identified one variant, rs4057749 (P = 5.39 × 10−9), within OR8B4 gene at an exome-wide significance level. The gene-based burden test of the exonic variants identified genome-wide significant associations in OR8B4 (PSKAT = 6.23 × 10−5, PSKAT Robust = 4.49 × 10−5), TRAPPC11 (PSKAT = 0.014, PSKAT Robust = 0.015), SBK3 (PSKAT = 0.020, PSKAT Robust = 0.025) and TNRC6B (PSKAT = 0.026, PSKAT Robust = 0.036). We identified multiple novel rare risk variants contributing to MDD in the individuals with lower PRS of MDD. The findings can help to broaden the genetic insights of the MDD pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The UK Biobank data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. We will return the derived data fields following UK Biobank policy; in due course, they will be available through the UK Biobank Access Management System.
Code availability
All scripts used to generate the SKAT analyses are available from the authors upon request.
References
James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–858.
Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–62.
Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Culverhouse RC, Saccone NL, Horton AC, Ma Y, Anstey KJ, Banaschewski T, et al. Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol Psychiatry. 2018;23:133–42.
Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888.
Bis JC, Jian X, Kunkle BW, Chen Y, Hamilton-Nelson KL, Bush WS, et al. Whole exome sequencing study identifies novel rare and common Alzheimer’s-Associated variants involved in immune response and transcriptional regulation. Mol Psychiatry. 2020;25:1859–75.
Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50:26–41.
Lu T, Zhou S, Wu H, Forgetta V, Greenwood CMT, Richards JB. Individuals with common diseases but with a low polygenic risk score could be prioritized for rare variant screening. Genet Med: Off J Am Coll Med Genet. 2021;23:508–15.
Bomba L, Walter K, Soranzo N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017;18:77.
Zhou D, Yu D, Scharf JM, Mathews CA, McGrath L, Cook E, et al. Contextualizing genetic risk score for disease screening and rare variant discovery. Nat Commun. 2021;12:4418.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9.
Szustakowski JD, Balasubramanian S, Kvikstad E, Khalid S, Bronson PG, Sasson A, et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat Genet. 2021;53:942–8.
Lin MF, Rodeh O, Penn J, Bai X, Reid JG, Krasheninina O, et al. GLnexus: joint variant calling for large cohort sequencing. bioRxiv 2018:343970.
Kroenke K, Spitzer RL, Williams JBW, Löwe B. The patient health questionnaire somatic, anxiety, and depressive symptom scales: a systematic review. Gen Hosp Psychiatry. 2010;32:345–59.
Euesden J, Lewis CM, Oreilly PF. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–68.
Lambert SA, Gil L, Jupp S, Ritchie SC, Xu Y, Buniello A, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat Genet. 2021;53:420–5.
Cai N, Revez JA, Adams MJ, Andlauer TFM, Breen G, Byrne EM, et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat Genet. 2020;52:437–47.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Bjørnland T, Bye A, Ryeng E, Wisløff U, Langaas M. Powerful extreme phenotype sampling designs and score tests for genetic association studies. Stat Med. 2018;37:4234–51.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e64.
Cirulli ET, White S, Read RW, Elhanan G, Metcalf WJ, Tanudjaja F, et al. Genome-wide rare variant analysis for thousands of phenotypes in over 70,000 exomes from two cohorts. Nat Commun. 2020;11:542.
Sun YV, Sung YJ, Tintle N, Ziegler A. Identification of genetic association of multiple rare variants using collapsing methods. Genet Epidemiol. 2011;35:S101–6.
Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–75.
Lee S, Fuchsberger C, Kim S, Scott L. An efficient resampling method for calibrating single and gene-based rare variant association analysis in case-control studies. Biostatistics. 2016;17:1–15.
Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–37.
Zhao Z, Bi W, Zhou W, VandeHaar P, Fritsche LG, Lee S. UK biobank whole-exome sequence binary phenome analysis with robust region-based rare-variant test. Am J Hum Genet. 2020;106:3–12.
Kalmbach DA, Schneider LD, Cheung J, Bertrand SJ, Kariharan T, Pack AI, et al. Genetic basis of chronotype in humans: insights from three landmark GWAS. Sleep. 2016;40.
Zhang Y, Emery P. GW182 controls Drosophila circadian behavior and PDF-receptor signaling. Neuron. 2013;78:152–65.
Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54.
Babbs C, Lloyd D, Pagnamenta AT, Twigg SR, Green J, McGowan SJ, et al. De novo and rare inherited mutations implicate the transcriptional coregulator TCF20/SPBP in autism spectrum disorder. J Med Genet. 2014;51:737–47.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Granadillo JL, A PAS, Guo H, Xia K, Angle B, Bontempo K, et al. Pathogenic variants in TNRC6B cause a genetic disorder characterised by developmental delay/intellectual disability and a spectrum of neurobehavioural phenotypes including autism and ADHD. J Med Genet. 2020;57:717–24.
Ackerman S, Schoenbrun S, Hudac C, Bernier R. Interactive effects of prenatal antidepressant exposure and likely gene disrupting mutations on the severity of autism spectrum disorder. J Autism Developmental Disord. 2017;47:3489–96.
Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci. 2004;101:2584–9.
Bögershausen N, Shahrzad N, Chong JX, von Kleist-Retzow JC, Stanga D, Li Y, et al. Recessive TRAPPC11 mutations cause a disease spectrum of limb girdle muscular dystrophy and myopathy with movement disorder and intellectual disability. Am J Hum Genet. 2013;93:181–90.
Koehler K, Milev MP, Prematilake K, Reschke F, Kutzner S, Jühlen R, et al. A novel TRAPPC11 mutation in two Turkish families associated with cerebral atrophy, global retardation, scoliosis, achalasia and alacrima. J Med Genet. 2017;54:176–85.
Matalonga L, Bravo M, Serra-Peinado C, García-Pelegrí E, Ugarteburu O, Vidal S, et al. Mutations in TRAPPC11 are associated with a congenital disorder of glycosylation. Hum Mutat. 2017;38:148–51.
Wetherill L, Lai D, Johnson EC, Anokhin A, Bauer L, Bucholz KK, et al. Genome-wide association study identifies loci associated with liability to alcohol and drug dependence that is associated with variability in reward-related ventral striatum activity in African- and European-Americans. Genes, Brain Behav. 2019;18:e12580.
MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45:D896–d901.
Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–9.
Lanktree MB, Hegele RA, Schork NJ, Spence JD. Extremes of unexplained variation as a phenotype. circulation: cardiovascular. Genetics. 2010;3:215–21.
Acknowledgements
This study was conducted using the UK Biobank Resource (Application 46478).
Funding
This study was supported by the National Natural Scientific Foundation of China (81922059).
Author information
Authors and Affiliations
Contributions
SC and FZ conceived and designed the study, and wrote the manuscript; SC and FZ collected the data and carried out the statistical analyses; BC, LL, XL, PM, YY, CP, JZ, CL, HZ, YC, ZZ, YW, and YJ made preparations for the manuscript at first.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Ethical approval of UK Biobank study was granted by the National Health Service National Research Ethics Service (reference 11/NW/0382).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cheng, S., Cheng, B., Liu, L. et al. Exome-wide screening identifies novel rare risk variants for major depression disorder. Mol Psychiatry 27, 3069–3074 (2022). https://doi.org/10.1038/s41380-022-01536-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01536-4
This article is cited by
-
Large-scale whole-exome sequencing of neuropsychiatric diseases and traits in 350,770 adults
Nature Human Behaviour (2024)
-
Integration of whole-exome sequencing and structural neuroimaging analysis in major depressive disorder: a joint study
Translational Psychiatry (2024)
-
Exome-wide association study of treatment-resistant depression suggests novel treatment targets
Scientific Reports (2023)
-
Exome-wide screening identifies novel rare risk variants for bone mineral density
Osteoporosis International (2023)
-
Examining the source of increased bipolar disorder and major depressive disorder common risk variation burden in multiplex schizophrenia families
Schizophrenia (2022)