Abstract
Human papillomavirus-negative keratinizing vulvar cancers typically harbor TP53 mutations as do their precursors, differentiated vulvar intraepithelial neoplasia. However, atypical verruciform proliferations are also associated with these malignancies and their pathogenesis is poorly understood. This study compared 11 atypical verruciform lesions, including atypical verruciform hyperplasia, vulvar acanthosis with altered differentiation, and verruciform lichen simplex chronicus, with 14 human papillomavirus-negative keratinizing squamous cell carcinomas. Extracted tissue DNA was subjected to targeted massively parallel sequencing of the exonic regions of 300 genes. Eight (73%) and six (55%) of eleven atypical verruciform lesions contained mutations in PIK3CA and ARID2, respectively. No TP53 mutations were identified. Eleven (79%) and five (36%) of fourteen keratinizing squamous cell carcinomas tested contained TP53 and CDKN2A mutations, respectively. Keratinizing squamous cell carcinomas displayed the majority of copy number variations with some variations (7p gain and 8p loss) shared by some cases in both groups. One patient developed atypical verruciform lesions with PIK3CA mutations followed by a keratinizing carcinoma with mutations in both PIK3CA and TP53. This study, for the first time segregates atypical verruciform lesions by virtue of a unique genotype (PIK3CA mutant/TP53 wild type) illustrating an example of progression to a TP53-mutated keratinizing carcinoma. The findings indicate that although PIK3CA mutations are found in <10% of vulvar squamous cell carcinomas, they may be specific for a particular pathway involving atypical verruciform lesions, which could function as either a direct precursor or a risk factor for vulvar squamous cell carcinoma. Given the presence of a molecular signature, we propose the term ‘differentiated exophytic vulvar intraepithelial lesion’ for this group. Whether they function as direct precursors to a less common form of squamous cell carcinoma will require further study, but carcinomas associated with these lesions might warrant testing for PIK3CA mutations to address this question.
Similar content being viewed by others
Main
Vulvar squamous cell carcinoma, a rare cancer with an annual incidence of 2 per 100 000 women in developed countries,1, 2, 3 typically derives from one of two pathophysiologic pathways. The first stems from human papillomavirus infection, which gives rise to classic (usual) vulvar intraepithelial neoplasia, the precursor lesion of basaloid, moderately to poorly differentiated squamous cell carcinomas.4, 5, 6, 7, 8 The second route is human papillomavirus-independent and is primarily driven by TP53 mutations.5, 6, 8, 9, 10 The known precursor lesions in this pathway, including lichen sclerosus, lichen simplex chronicus, and differentiated vulvar intraepithelial neoplasia, give rise to keratinizing squamous cell carcinoma.5, 6, 8, 9, 10
However, approximately one-third of vulvar squamous cell carcinomas, primarily those that are human papillomavirus-negative, are not associated with a conventional precursor lesion (eg, differentiated vulvar intraepithelial neoplasia, classic vulvar intraepithelial neoplasia) at the time of diagnosis.11, 12 At our institution, we have noted an association between human papillomavirus-negative keratinizing squamous cell carcinoma and a variety of atypical verruciform lesions, many of which have previously defied classification and do not meet traditional criteria for vulvar intraepithelial neoplasia or other known precursor lesions. Such lesions have been previously defined as vulvar acanthosis with altered differentiation,12 verruciform lichen simplex chronicus, or atypical verruciform hyperplasia. A hallmark of this group is acanthosis or verruciform growth pattern with abnormal keratinocyte differentiation in the absence of conspicuous nuclear atypia. Given the absence of atypia, these lesions have yet to be universally classified as precancers, resulting in non-uniform classification and clinical management. However, given their observed association with keratinizing squamous cell carcinoma, it is possible that these lesions represent a either a distinct group of precursor lesions or a risk factor for keratinizing carcinoma.12
We designed a study to determine if atypical verruciform lesions contain molecular perturbations and if so, are they linked to keratinizing squamous cell carcinomas or do they reflect an alternate pathogenesis. Given that atypical verruciform lesions lack basal nuclear atypia and probably do not harbor TP53 mutations, study of these lesions might unearth additional gene mutations or pathways that either contribute to the genesis of keratinizing squamous cell carcinomas or are emblematic of underlying risk for malignancy.
Materials and Methods
Case Selection and Criteria for Atypical Verruciform Lesions
This project was approved by the Institutional Review Board at Brigham and Women's Hospital. Cases diagnosed as atypical verruciform lesions or non-human papillomavirus-associated keratinizing squamous cell carcinomas were retrieved from the surgical pathology files of the Division of Women's and Perinatal Pathology, Brigham and Women's Hospital, Boston, Massachusetts. For all cases, the hematoxylin and eosin slides were reviewed for confirmation of the diagnosis by three pathologists (BH, CC, and JW).
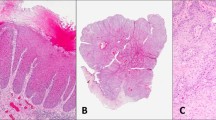
Atypical verruciform lesions were defined by the following criteria: (1) they were exophytic with prominent acanthosis or verruciform architecture; (2) they lacked the histomorphologic features characteristic of human papillomavirus-related lesions (eg, koilocytotic atypia or atypia in all layers of the epithelium, nuclear hyperchromasia, high nuclear to cytoplasmic ratio, and pleomorphism);13, 14 3) they lacked sufficient basal atypia to warrant a diagnosis of differentiated vulvar intraepithelial neoplasia (Figure 1). Cases in the atypical verruciform lesion group thus included lesions with ambiguous features that fell into the realm of vulvar acanthosis with altered differentiation, including lesions bordering on verrucous carcinoma, verruciform lichen simplex chronicus, or atypical verruciform hyperplasia. Unequivocal verrucous carcinoma cases were excluded, and only those cases that were equivocal for invasion (ie, lacked clear invasion through an expansile blunt interface) were included in this study. All such cases were designated atypical verruciform lesions for the purposes of analyses.
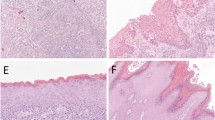
(a) A PIK3CA, CDKN2A, and TP53-mutated invasive keratinizing squamous cell carcinoma, well-differentiated (case 1) arising in association with an atypical verruciform lesion (b) with mild basal atypia (c) insufficient for a diagnosis of differentiated vulvar intraepithelial neoplasia. (d) A PIK3CA mutated case of an atypical verruciform lesion (case 15) demonstrating exophytic growth, minimal basal atypia (e) and an absence of human papillomavirus-associated histomorphologic changes (f). (g) A PIK3CA mutated verruciform, exophytic example of an atypical verruciform lesion (case 17) with minimal basal atypia (h) and no evidence of human papillomavirus-related changes (i). (j) A PIK3CA mutated exophytic, verruciform atypical verruciform lesion (case 19) with absent human papillomavirus-related changes (k) and minimal basal atypia (l). (m) An example of an atypical verruciform lesion (case 23) with no mutations found through massively parallel sequencing with exophytic growth and minimal basal atypia (n).
Cases of keratinizing squamous cell carcinoma were only included if they were well-to-moderately differentiated and lacked basaloid features suggestive of human papillomavirus infection. When documented in the pathology report, precursor lesions, either apparent simultaneously at the time of invasive squamous cell carcinoma or present in a prior biopsy, were recorded. In addition, when material was available, cases of keratinizing squamous cell carcinoma that arose in association with atypical verruciform lesions were included.
After hematoxylin and eosin review, only cases with sufficient residual material for molecular profiling were included (ie, block with adequate remaining tissue or at least five unstained slides). A total of 25 cases were included in the study, including 11 atypical verruciform lesions, and 14 non-human papillomavirus-related keratinizing squamous cell carcinomas.
Immunohistochemistry
When available, p53 immunohistochemical staining originally obtained during the work-up of the atypical verruciform lesion cases was reviewed. If a p53 immunostain had not been performed at the time of diagnosis, cases were stained immunohistochemically for p53 via the Envision Plus detection system (Dako, Carpinteria, CA, USA). The following antibody and dilution was applied: p53 (1:1200). Antigen retrieval entailed heat-induced epitope retrieval in 10 mM citrate buffer with a pressure cooker. Appropriate positive and negative controls were used throughout.
Molecular Profiling
Next-generation sequencing of the lesions was performed. In brief, DNA was isolated from unstained formalin-fixed paraffin-embedded tissue slides (Qiagen) or by coring paraffin-embedded blocks and quantified (PicoGreen Eugene, OR). Samples meeting a minimum criterion of 50 ng/μl of DNA proceeded to library preparation. Hybrid capture libraries were prepared based on established protocols.15, 16 Sheared DNA was hybridized to a set of custom capture probes (Agilent SureSelect), which target the complete exonic regions of 300 cancer genes including TP53, HRAS, PIK3CA, and EGFR (complete list of genes included in Supplementary Table 1). Sequencing was performed using Illumina HiSeq 2500. Data was analyzed by an internally-developed bioinformatics pipeline, composed of internally-developed algorithms and reconfigured publically-available tools for detection of single nucleotide variants and indels (MuTect, GATK). Tumor tissues were tested without a paired normal. Therefore, any single nucleotide variant present at >0.1% in Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, USA (URL: http://evs.gs.washington.edu/EVS/, accessed 5/30/2016) was filtered and removed from factor analysis. Variants also present in the Catalogue of Somatic Mutations in Cancer (COSMIC; cancer.sanger.ac.uk) were rescued for manual review.
Results
Characteristics of the Study Population
Eleven atypical verruciform lesions from nine patients and fourteen well-to-moderately differentiated-keratinizing squamous cell carcinomas from fourteen patients were retrieved for review. Two cases of keratinizing squamous cell carcinoma arose in patients with simultaneously profiled atypical verruciform lesions. For the keratinizing squamous cell carcinoma cases, patient ages, diagnoses, and precursor lesions are documented in Table 1. For the atypical verruciform lesion cases, patient ages, initial diagnoses as well as diagnosis after detailed pathologic review, and outcomes are outlined in Table 2.
Overview of Single Nucleotide Variations and Indels in Atypical Verruciform Lesions and Keratinizing Squamous Cell Carcinomas
A total of 43 single nucleotide variants in 29 genes were identified across the atypical verruciform lesions, including 27 missense, 6 nonsense, 4 frameshift, 5 splice site, and 1 initiator codon mutations (Figure 2). Of the missense mutations, 12 had previously been reported in other tumor types in COSMIC (cancer.sanger.ac.uk). The mean number of single nucleotide variants per atypical verruciform lesion was 4 (range 0–12, median=2).
Recurrent single nucleotide and copy number variations in vulvar atypical verruciform lesions and vulvar keratinizing squamous cell carcinomas. Each row represents a gene and each column represents a case. Two cases of keratinizing squamous cell carcinoma arising in association with atypical verruciform lesions are included (cases 24 and 25). Case 24 represents the squamous cell carcinoma arising in association with cases 14, 15, and 16. Case 25 arose in association with case 19.
Overall, 49 single nucleotide variants in 28 genes were identified across the keratinizing squamous cell carcinomas, including 32 missense, 7 nonsense, 5 frameshift, 3 splice site, and 1 initiator codon mutations as well as 1 inframe deletion. Sixteen of the missense mutations had been previously reported in other tumor types in COSMIC. The mean number of single nucleotide variants per keratinizing squamous cell carcinoma was 4 (range 1–21, median=2).
A complete list of all single nucleotide variants identified across all cases is available in Supplementary Table 2.
Overview of Copy Number Variations in Atypical Verruciform Lesions and Keratinizing Squamous Cell Carcinomas
The majority of copy number variations occurred in keratinizing squamous cell carcinomas and included whole chromosome, arm level alterations, and few gene level copy number variations (Figure 2). Three atypical verruciform lesions displayed copy number variations. All keratinizing squamous cell carcinomas demonstrated copy number variations, with three cases demonstrating gene level copy number variations in addition to arm level alterations.
TP53 Mutations are not Present in Atypical Verruciform Lesions
Mutations in TP53 were extremely common in the keratinizing squamous cell carcinoma cohort, with 13 identified mutations (10 missense, 1 inframe deletion, 1 nonsense, and 1 slice site mutation) present in 11 of 14 cases (79%). Two missense mutations occurred in two cases (Table 1; cases 6 and 24), suggesting biallelic inactivation. In addition, copy number loss of TP53/17p was seen in 6 of the 11 TP53-mutated keratinizing squamous cell carcinomas (55%) (see below), providing evidence of biallelic inactivation in these cases. Notably, none of the atypical verruciform lesions harbored mutations in TP53.
Activating PIK3CA Mutations are Common in Atypical Verruciform Lesions but not in Keratinizing Squamous Cell Carcinoma
Eight (73%) of the atypical verruciform lesions demonstrated missense mutations in PIK3CA (5 E545K, 1 H1047R, 1 N515S, 1 R519T, 1 E542K, and 1 D549H), the majority of which were well-documented activating mutations. Of these cases, four of the atypical verruciform lesions harboring E545K or H1047R mutations were associated with subsequent invasive keratinizing squamous cell carcinoma. Two of the keratinizing squamous cell carcinoma cases demonstrated mutations in PIK3CA (E453K and E545K; Table 1, cases 1 and 24, respectively). In case 1, the patient had presented prior to her cancer diagnosis with an ill-defined verruciform vulvar lesion with parakeratosis and focal squamous atypia in addition to lichen sclerosus. However, material from this lesion was unavailable for molecular testing. Case 24 is discussed below.
Recurrent Atypical Verruciform Lesions and Squamous Cell Carcinoma (Case 24) in the Same Patient
Three atypical verruciform lesions from the same patient (cases 14, 15, and 16; Figure 2) were included in the study. The patient was initially diagnosed with verruciform lichen simplex chronicus (case 16), followed by verruciform-differentiated vulvar intraepithelial neoplasia and atypical verruciform hyperplasia (case 14), atypical verruciform hyperplasia (case 15), and invasive well-to-moderately differentiated squamous cell carcinoma, with each lesion occurring approximately one year apart. All three of the atypical verruciform lesions displayed PIK3CA, ARID2, and ROS1 mutations. The carcinoma (Table 1; case 24) harbored mutations in both PIK3CA and TP53 (Figure 2).
A second case of keratinizing squamous cell carcinoma (Table 1; case 25) from a patient with an atypical verruciform lesion (Table 2; case 19) was also profiled. The carcinoma lacked mutations in PIK3CA and TP53 suggesting that it was not directly related to the atypical verruciform lesion.
HRAS Mutations in Atypical Verruciform Lesions and Keratinizing Squamous Cell Carcinomas
Two (18%) of the atypical verruciform lesions demonstrated missense mutations in HRAS, including 1 G12S and 1 G13V, both of which are activating mutations. Follow up available for the G12S mutant revealed that the patient subsequently developed keratinizing squamous cell carcinoma (Table 2; case 14). A single HRAS mutation was identified in the keratinizing squamous cell carcinoma group (G13D) (Table 1; case 5).
ARID2 Mutations in Atypical Verruciform Lesions and Keratinizing Squamous Cell Carcinomas
Six mutations in ARID2 (2 frameshift, 3 splice site, and 1 initiator codon mutation) were observed in four atypical verruciform lesions from two patients (2/9, 22%). The two frameshift mutations (D196Rfs, D1459Ffs) occurred in the same case (Table 2; case 14), suggesting possible biallelic inactivation. This lesion occurred in a patient who subsequently developed keratinizing squamous cell carcinoma. Only one mutation in ARID2 was observed in the keratinizing squamous cell carcinoma group (M1346Cfs) (Table 1; case 5).
CDKN2A Mutations are Common in Keratinizing Squamous Cell Carcinomas
Five mutations in CDKN2A (3 nonsense, 1 initiator codon, and 1 frameshift) were observed in four cases (4/14, 29%) of keratinizing squamous cell carcinoma. A single case (Table 1; case 4) demonstrated both a nonsense (R58*) and frameshift (A76Rfs) mutation, suggestive of biallelic inactivation. An additional case of keratinizing squamous cell carcinoma demonstrated copy number loss of CDKN2A/9p, also suggestive of biallelic inactivation. One nonsense mutation (R58*) was observed in the atypical verruciform lesion group, a case in which the patient subsequently developed keratinizing squamous cell carcinoma (Table 2; case 13).
Recurrent Copy Number Variations in Atypical Verruciform Lesions and Keratinizing Squamous Cell Carcinoma
The majority of the chromosomal and arm level gains/losses were identified in keratinizing squamous cell carcinoma (Figure 2). Seven keratinizing squamous cell carcinomas demonstrated 3p loss, with one of the cases demonstrating only partial loss. Eight keratinizing squamous cell carcinomas demonstrated 3q gain, with one case only demonstrating partial gain. Five keratinizing squamous cell carcinomas demonstrated 7p gain. 8p loss was seen in 8 keratinizing squamous cell carcinomas, 8q gain was seen in 9 keratinizing squamous cell carcinomas, and 17p loss was seen in 7 keratinizing squamous cell carcinomas.
The atypical verruciform lesions demonstrated fewer copy number variations. One case (Table 1; case 13), in which the patient subsequently developed keratinizing squamous cell carcinoma, demonstrated a 7p gain and 8p loss. A second case with 7p gain (Table 2; case 20) developed keratinizing squamous cell carcinoma. One additional lesion (Table 2; cases 18) demonstrated 7p gain. No follow up was available for this patient.
High gene level amplification of CCND1 and EGFR was identified in keratinizing squamous cell carcinomas (n=2 and 1, respectively). The two cases demonstrating high gene level amplification of CCND1 had an estimated 18.5 and 19.9 copies of CCND1, respectively. The case demonstrating EGFR amplification had an estimated 22.4 copies of EGFR. No structural variants were identified.
Discussion
The pathogenesis of vulvar cancer is currently dichotomous. HPV positive tumors, which develop from usual-type vulvar intraepithelial neoplasia, are defined by high-risk HPV infection—usually HPV 16. The HPV-negative pathway is best known for originating in differentiated vulvar intraepithelial neoplasia, which is associated with TP53 mutations. Thus, apart from clinical and histologic parameters, HPV and TP53 are the two prevalent biomarkers for separating these two entities. However, a significant, albeit small, subset of vulvar carcinomas exhibits neither HPV infection nor mutations in TP53. Some of these carcinomas have been associated with atypical verruciform squamous lesions that do not fulfill the histologic criteria for either usual or differentiated high-grade squamous intraepithelial lesion. The term ‘atypical verruciform hyperplasia’ is often applied in such occasions, but the biologic characteristics of this entity and its role in vulvar squamous carcinogenesis have remained unclear. Early studies alluded to allelic imbalance in these lesions.17 However, given the explosion of molecular information enabled by sophisticated high throughput genomic sequencing, there is now the opportunity to classify these and other disorders from a new perspective. In this study we evaluated 11 vulvar atypical verruciform lesions and 14 vulvar keratinizing squamous cell carcinomas through targeted massively parallel sequencing (OncoPanel). The goal was to initiate the discovery of a framework of alterations upon which the morphologic groupings can be superimposed and examined. The results have shown that atypical verruciform lesions might be separated from these other forms of vulvar squamous neoplasia by unique molecular biomarkers.
The first finding in this study fundamental to the separation of atypical verruciform lesions from either differentiated vulvar intraepithelial neoplasia or keratinizing squamous cell carcinoma was the difference in TP53 mutation status. Some verruciform lesions exhibited basal cell atypia and some stained positively for p53; nevertheless, none of these lesions harbored a TP53 mutation. In contrast, 79% of our keratinizing squamous cell carcinoma cases were TP53 mutation positive. These results suggest that atypical verruciform lesions, whereas heterogeneous in appearance, should be separated from differentiated VIN as currently defined.
The second and pivotal finding for discriminating atypical verruciform lesions from the majority of keratinizing squamous cell carcinomas pertained to PIK3CA. Activating PIK3CA mutations were identified in 73% of verruciform lesions in contrast to 14% (2 of 14) of cases of keratinizing carcinoma. Noteworthy was that the two keratinizing carcinomas with both PIK3CA and TP53 mutations arose within fields of atypical verruciform lesions (cases 1 and 24). Activating PIK3CA mutations have been documented previously in carcinomas of multiple sites. However, only one study so far has identified such mutations in vulvar carcinoma, with an incidence of only 8% in human papillomavirus-negative lesions.18 The incidence is much higher in our atypical verruciform lesion cohort, with 73% of lesions demonstrating activating mutations in PIK3CA. Although we cannot exclude that other mutations may ultimately drive carcinogenesis in these lesions, especially as this study lacks intra-patient atypical verruciform lesion/keratinizing squamous cell carcinoma cases, the ubiquity of PIK3CA mutations in the absence of TP53 mutations suggests PIK3CA mutations may be the common driver mutation in many atypical verruciform lesions. This has potential clinical impact as PI3K inhibitors could theoretically be used as a treatment option in such cases if they advance to invasive squamous cell carcinoma.
The absence of TP53 mutations, combined with the high frequency of PIK3CA mutations in atypical verruciform lesions is in keeping with their separation from both classic and differentiated vulvar intraepithelial neoplasia. Furthermore, if we assume that PIK3CA mutations have a role in the development of atypical verruciform lesions and their progression to keratinizing squamous cell carcinoma pathway, then it is possible that the PI3K/AKT/mTOR pathway represents a third, albeit less common pathway to vulvar carcinoma. Given their unique morphologic and genetic characteristics relative to differentiated and usual vulvar intraepithelial neoplasia, atypical verruciform lesions deserve a separate designation. Thus, we propose the provisional term ‘differentiated exophytic vulvar intraepithelial lesion’ for the atypical verruciform lesion group and define this group as manifesting with (1) verruciform morphology, (2) abnormalities in keratinocyte differentiation, (3) absence of conspicuous basal atypia, (4) absence of TP53 mutations, and (5) a high frequency of PIK3CA mutations. Their association with keratinizing squamous cell carcinoma, either as a concurrent or preceding phenomenon, argues for a management scheme wherein differentiated exophytic vulvar intraepithelial lesion is viewed as a risk factor for keratinizing squamous cell carcinoma, whereas their role as an immediate precursor is under further study.
Parenthetically, the reason for this approach is that multiple genetic alterations can be found in the vulvar mucosa of women with keratinizing squamous cell carcinoma and pinpointing the precise precursor can be difficult. In cases 1 and 24, a differentiated exophytic vulvar intraepithelial lesion was associated with keratinizing squamous cell carcinoma, the latter with mutations in both TP53 and PIK3CA. However, case 1 was complicated by the presence of LS, which can also be associated with TP53-mutations.19 Thus, we emphasize that this study does not prove a direct link between all differentiated exophytic vulvar intraepithelial lesions and keratinizing squamous cell carcinomas but it underscores the fact that important mutations can occur in vulvar lesions that are known to increase the risk of developing subsequent keratinizing squamous cell carcinoma. This is similar to prior observations by this group mapping allelic loss in the vulvar mucosa of women with keratinizing squamous cell carcinoma.20, 17 Whether differentiated exophytic vulvar intraepithelial lesions acquire TP53 mutations and proceed directly to keratinizing squamous cell carcinoma, and whether a subset of keratinizing squamous cell carcinomas without TP53 mutations arose from differentiated exophytic vulvar intraepithelial lesions remains to be proven by more detailed studies analyzing multiple lesions in the same subject.
Three additional gene mutations of interest were HRAS, ARID2, and CDKN2A. HRAS mutations have been previously found in up to 11% of human papillomavirus-negative vulvar keratinizing squamous cell carcinoma, and in fact, are associated with worse prognoses, especially when coupled with a TP53 mutation.18 One case in our keratinizing squamous cell carcinoma cohort (7%) demonstrated an HRAS mutation, and two cases of differentiated exophytic vulvar intraepithelial lesion (18%) demonstrated HRAS mutations, suggesting a possible pathogenic role in the differentiated exophytic vulvar intraepithelial lesion to keratinizing squamous cell carcinoma transition. Such findings may have clinical relevance as dual blockade of RAS and PIK3CA by combined therapy with MEK and PI3K inhibitors may lead to better clinical outcomes for patients with either simultaneous mutations or a single mutation in either gene.18, 21, 22 In addition, one case of keratinizing squamous cell carcinoma demonstrated EGFR amplification, which like the PIK3CA mutated differentiated exophytic vulvar intraepithelial lesions, may be treated with targeted therapy. Other interesting findings in this study include the first documented ARID2 mutation in vulvar squamous cell carcinoma and confirmation of previously described CDKN2A mutations in vulvar squamous cell carcinoma.18 The copy number variation findings in the keratinizing squamous cell carcinoma cohort are in keeping with those previously documented in vulvar squamous cell carcinoma.23
References
Judson PL, Habermann EB, Baxter NN et al, Trends in the incidence of invasive and in situ vulvar carcinoma. Obstet Gynecol 2006;107:1018–1022.
Schuurman MS, van den Einden LCG, Massuger LFAG et al, Trends in incidence and survival of Dutch women with vulvar squamous cell carcinoma. Eur J Cancer 2013;49:3872–3880.
van de Nieuwenhof HP, LFAG Massuger, van der Avoort IAM et al, Vulvar squamous cell carcinoma development after diagnosis of VIN increases with age. Eur J Cancer 2009;45:851–856.
Kurman RJ, Toki T, Schiffman MH. . Basaloid and warty carcinomas of the vulva. Distinctive types of squamous cell carcinoma frequently associated with human papillomaviruses. Am J Surg Pathol 1993;17:133–145.
McCluggage WG. . Recent developments in vulvovaginal pathology. Histopathology 2009;54:156–173.
Del Pino M, Rodriguez-Carunchio L, Ordi J. . Pathways of vulvar intraepithelial neoplasia and squamous cell carcinoma. Histopathology 2013;62:161–175.
De Vuyst H, Clifford GM, Nascimento MC et al, Prevalence and type distribution of human papillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: a meta-analysis. Int J Cancer 2009;124:1626–1636.
Fox H, Wells M. . Recent advances in the pathology of the vulva. Histopathology 2003;42:209–216.
Yang B, Hart WR. . Vulvar intraepithelial neoplasia of the simplex (differentiated) type: a clinicopathologic study including analysis of HPV and p53 expression. Am J Surg Pathol 2000;24:429–441.
Roma AA, Hart WR. . Progression of simplex (differentiated) vulvar intraepithelial neoplasia to invasive squamous cell carcinoma: a prospective case study confirming its precursor role in the pathogenesis of vulvar cancer. Int J Gynecol Pathol 2007;26:248–253.
Leibowitch M, Neill S, Pelisse M et al, The epithelial changes associated with squamous cell carcinoma of the vulva: a review of the clinical, histological and viral findings in 78 women. Br J Obstet Gynaecol 1990;97:1135–1139.
Nascimento AF, Granter SR, Cviko A et al, Vulvar acanthosis with altered differentiation: a precursor to verrucous carcinoma? Am J Surg Pathol 2004;28:638–643.
Reyes MC, Cooper K. . An update on vulvar intraepithelial neoplasia: terminology and a practical approach to diagnosis. J Clin Pathol 2014;67:290–294.
Hart WR. . Vulvar intraepithelial neoplasia: historical aspects and current status. Int J Gynecol Pathol 2001;20:16–30.
Abo RP, Ducar M, Garcia EP et al, BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res 2014;43:e19.
Wagle N, Berger MF, Davis MJ et al, High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2012;2:82–93.
Pinto AP, Lin MC, Sheets EE et al, Allelic imbalance in lichen sclerosus, hyperplasia, and intraepithelial neoplasia of the vulva. Gynecol Oncol 2000;77:171–176.
Trietsch MD, Spaans VM, ter Haar NT et al, CDKN2A(p16) and HRAS are frequently mutated in vulvar squamous cell carcinoma. Gynecol Oncol 2014;135:149–155.
Rolfe KJ, MacLean a B, Crow JC et al, TP53 mutations in vulval lichen sclerosus adjacent to squamous cell carcinoma of the vulva. Br J Cancer 2003;89:2249–2253.
Pinto AP, Lin MC, Mutter GL et al, Allelic loss in human papillomavirus-positive and -negative vulvar squamous cell carcinomas. Am J Pathol 1999;154:1009–1015.
Castellano E, Downward J . RAS interaction with PI3K: more than just another effector pathway. Genes Cancer 2011;2:261–274.
Roper J, Sinnamon MJ, Coffee EM et al, Combination PI3K/MEK inhibition promotes tumor apoptosis and regression in PIK3CA wild-type, KRAS mutant colorectal cancer. Cancer Lett 2014;347:204–211.
Trietsch MD, Nooij LS, Gaarenstroom KN et al, Genetic and epigenetic changes in vulvar squamous cell carcinoma and its precursor lesions: a review of the current literature. Gynecol Oncol 2015;136:143–157.
Acknowledgements
Portions of this study were presented in poster format at the United States and Canadian Academy of Pathology 2016 Annual Meeting, held in Seattle, Washington on March 12, 2016. We are grateful to the Division of Gynecologic Oncology (Michael Muto, Colleen Feltmate, and Donald Goldstein) and to Dr Jerome Federschneider for their support and contribution of this case material in this study. We also acknowledge the Center for Advanced Molecular Diagnostics at Brigham and Women's Hospital and the Dana-Farber Cancer Institute for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Watkins, J., Howitt, B., Horowitz, N. et al. Differentiated exophytic vulvar intraepithelial lesions are genetically distinct from keratinizing squamous cell carcinomas and contain mutations in PIK3CA. Mod Pathol 30, 448–458 (2017). https://doi.org/10.1038/modpathol.2016.187
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.187
This article is cited by
-
HPV-independent, p53-wild-type vulvar intraepithelial neoplasia: a review of nomenclature and the journey to characterize verruciform and acanthotic precursor lesions of the vulva
Modern Pathology (2022)
-
Molecular landscape of vulvovaginal squamous cell carcinoma: new insights into molecular mechanisms of HPV-associated and HPV-independent squamous cell carcinoma
Modern Pathology (2022)
-
Molecular characterization of invasive and in situ squamous neoplasia of the vulva and implications for morphologic diagnosis and outcome
Modern Pathology (2021)
-
CDKN2A inhibits cell proliferation and invasion in cervical cancer through LDHA-mediated AKT/mTOR pathway
Clinical and Translational Oncology (2021)
-
Differentiated exophytic vulvar intraepithelial lesion: Clinicopathologic and molecular analysis documenting its relationship with verrucous carcinoma of the vulva
Modern Pathology (2020)