Abstract
The epidermal growth factor receptor is overexpressed in 70–90% of cervical cancers. Previously, we have shown that epidermal growth factor receptor overexpression independently predicts poor prognosis in cervical cancer patients, which makes it a potential therapeutic target. The aim of this study was to systematically analyze the molecular mechanism leading to epidermal growth factor receptor overexpression in cervical cancer. All experiments were performed on archival paraffin-embedded material. In 166 cervical cancer patients, cytoplasmic, membrane and phosphorylated epidermal growth factor receptor protein expression were studied in association with patient survival. Membrane epidermal growth factor receptor overexpression was associated with poor disease-specific survival (P=0.027). This association was particularly present in human papillomavirus 16-positive patients (P=0.029). We analyzed whether epidermal growth factor receptor overexpression was caused by gene amplification using fluorescence in situ hybridization. Epidermal growth factor receptor gene copy number was linked to chromosome 7 ploidy, as no gene amplification could be detected when corrected for chromosome 7 centromeric signals. Chromosome 7 aneuploidy was associated with membrane epidermal growth factor receptor overexpression (P=0.013). Additional mutation analysis was performed by sequencing pure, flow-sorted tumor cells, but no mutations were detected. Furthermore, human papillomavirus 16 E5 and E6 oncogene mRNA expression was measured, using quantitative real-time polymerase chain reaction, to determine the association between the human papillomavirus and epidermal growth factor receptor overexpression. High human papillomavirus 16 E5 and E6 mRNA expression were associated with decreased survival (P=0.045 and 0.047, respectively). High human papillomavirus 16 E6 mRNA expression was associated with membrane epidermal growth factor receptor overexpression (P=0.013). This is the first study performed on cancer patient material showing that chromosome 7 aneuploidy and high human papillomavirus 16 E6 mRNA expression lead to membrane epidermal growth factor receptor overexpression in cervical cancer.
Similar content being viewed by others
Main
Cervical cancer is the second most common cancer among women worldwide.1 Radical surgery and radiotherapy often provide satisfactory results for early-stage cervical cancer, but effective treatment options for patients with locally advanced disease are limited. With 5-year overall survival rates of ∼70% reported for chemo-/radiotherapy-treated patients, treatment response rates offer room for improvement.2, 3 The epidermal growth factor receptor (EGFR) is currently being explored as a therapeutic target in cervical cancer, as it is overexpressed in 70–90% of the cases.4, 5, 6 Encoded by the c-ErbB1 proto-oncogene on chromosome 7p, the EGFR is a member of the ErbB family of receptor tyrosine kinases. Activation of EGFR leads to cell growth, differentiation, resistance to apoptosis, cell cycle progression and angiogenesis.7 EGFR-based therapy is now being used for head and neck squamous cell carcinoma, colorectal carcinoma and non-small-cell lung carcinoma.8, 9, 10, 11 However, not all patients responded equally and knowledge of the mechanism of EGFR overexpression made it possible to select patients likely to benefit from EGFR-based therapy. Biomarkers predictive of response to EGFR therapy include gene amplification and activating mutations in EGFR.7, 12, 13 In cervical cancer, the mechanism leading to EGFR overexpression has not been clarified, even though EGFR-based therapy trials are currently being initiated.14
Previously, we have shown that EGFR overexpression is an independent predictor for poor prognosis in cervical cancer.15 More recently, EGFR overexpression has also been reported to be related to poor response to chemoradiation.16 Studies investigating the mechanism of EGFR overexpression in cervical cancer showed no EGFR mutations or gene amplifications, but sample sizes were small and EGFR protein expression was not determined.17, 18
The human papillomavirus (HPV) plays a central role in the carcinogenesis of cervical cancer. Expression of the E6 and E7 HPV oncogenes leads to the degradation and suppression of function of the p53 and retinoblastoma tumor suppressor proteins.19, 20 Although HPV oncogene expression has been associated with EGFR overexpression in vitro,21, 22, 23 studies substantiating these findings in cervical cancer patients have not been performed.
The aim of this study was to systematically investigate the molecular mechanisms leading to EGFR overexpression in cervical cancer. Therefore, we conducted a comprehensive study in a cohort of 166 cervical cancer patients. First, we examined cytoplasmic and membrane EGFR protein expression, as well as phosphorylated EGFR (pEGFR) expression, in association with disease-specific survival. Second, we analyzed whether EGFR overexpression is related to EGFR gene copy number or mutations in EGFR in the same patient cohort. Finally, we determined HPV E5 and E6 mRNA expression to assess the effect of these HPV oncogenes on EGFR overexpression in cervical cancer patients.
Materials and methods
Study Population
Formalin-fixed, paraffin-embedded tumor material from 166 cervical cancer patients who underwent radical hysterectomy with lymphadenectomy between 1985 and 2005 was retrieved from the archives of the Department of Pathology, Leiden University Medical Center, Leiden, the Netherlands. Patients had not received radio- or chemotherapy before surgery. Clinical and follow-up data were taken from patient medical records. Tumors were HPV-typed by general primer polymerase chain reaction (PCR) and sequencing, as described by Koopman et al.24 In short, HPV detection was performed by PCR using CP-I/II, GP5+/6+ and MY09/MY11 consensus primers. Samples that were found to be positive for HPV were subsequently sequenced to determine the HPV genotype. This study was approved by the Institutional Review Board according to the Dutch Federation of Medical Research Associations.
EGFR Protein Expression Analysis
Four-μm paraffin-embedded tissue sections were deparaffinized and rehydrated. Endogenous peroxidase was blocked with 0.3% hydrogen peroxide (H2O2) for 20 min. Antigen retrieval was performed in 0.4% pepsin in 0.01 M hydrochloric acid (HCl) (pH=2.0, 30 min, 37 °C, water bath) and 10 mM/0.5 mM Tris/ethylene glycol-tetraacetic acid (pH=9.0, 15 min, microwave oven) for EGFR and pEGFR, respectively. Slides were incubated overnight at room temperature with anti-EGFR (1:40, clone 31G7; Invitrogen, Breda, the Netherlands) diluted in PBS containing 1% bovine serum albumin (BSA) and overnight at 4 °C with anti-pEGFR (tyrosine 1173, 1:200, clone 53A5; Cell Signaling, Danvers, MA, USA) diluted in PBS/1% BSA with 5% normal goat serum. After washing with PBS, the tissue sections were incubated for 30 min with PowerVision-Poly/HRP (Immunologic, Duiven, the Netherlands). Immunoreactions were visualized using 0.5% 3.3′-diamino-benzidine-tetrahydrochloryde (DAB) and 0.002% H2O2 in Tris-HCl for EGFR and using a DAB+ chromogen (Dako, Heverlee, Belgium) for pEGFR. The sections were then counterstained with hematoxylin.
The intensity of both cytoplasmic/membrane staining and membrane staining of the EGFR (hereafter referred to as EGFR and mEGFR staining, respectively) were scored as negative (0), weak (1), moderate (2) or strong (3). Strong immunoreactivity was defined as complete membrane staining, producing a thick outline of the cell. Staining of the pEGFR was scored as either negative or positive membrane staining.
Gene Copy Number Analysis
Fluorescence in situ hybridization (FISH) on a tissue array containing 103 cervical cancer samples was performed as described previously.25 In short, the tissue array slide was incubated in Tris/EDTA buffer (pH=9.0) in a pressure cooker for 12.5 min. RNAse treatment was performed for 1 h at 37 °C (1 μg/ml 2 × SSC), followed by incubation with 0.1% pepsin in H2O (pH=2, 15 min, 37 °C). A chromosome 7 centromeric region probe (p7t1 alphoid satellite probe)26 was labelled with biotin by standard nick translation. For EGFR, PAC clone RP5-1091E12 was selected from the Ensembl Genome Browser (http://www.ensembl.org/Homo_sapiens/Location/Overview?r=7:54898372–55398371) and was labelled with digoxigenin-12-dUTP (Roche, Basel, Switzerland) by standard nick translation. The hybridization solution contained 50% formamide, 10% dextran sulfate, 50 mM sodium phosphate (pH=7.0), 2 × SCC, 3 ng/μl of each probe and a 50-fold excess of human Cot-1 DNA (BRL-Life Technologies, Rockville, MD, USA). After applying the probe mix, the slide was heated at 80 °C for 12 min and incubated overnight in a moist chamber at 37 °C. Immunodetection was performed as described previously.25 Images were captured using a COHU 4910 series monochrome CCD camera (COHU, San Diego, CA, USA) attached to a DM fluorescence microscope (Leica, Wetzlar, Germany) equipped with a PL Fluotar 100 × , NA 1.30–0.60 objective and I3 and N2.1 filters (Leica) and Leica QFISH software (Leica Imaging Systems, Cambridge, UK).
Mutation Analysis
DNA samples of 96 cervical cancer patients randomly selected from the study population were used. DNA was isolated from pure tumor cells, which were isolated from formalin-fixed, paraffin-embedded tumor specimens, using fluorescence-activated cell sorting, as described previously by Corver et al.27 DNA was amplified in a 10 μl reaction containing 3 μl DNA solution (approximately 500 cells per μl), 5 μl IQ mix, 0.5 μl primer and 1.5 μl H2O. Primer sequences for EGFR exons 18, 19, 20 and 2117 are shown in the Supplementary information available at Modern Pathology's website. Amplification conditions: 5 min initial denaturation at 95 °C, 40 cycles of 15 s at 95 °C, 15 s at 55 °C and 15 s at 72 °C, followed by 5 min extension at 72 °C. Sequencing was performed by the Leiden Genome Technology Centre (LGTC), Leiden, the Netherlands. The Mutation Surveyor program (SoftGenetics LLC, State College, PA, USA) was used for sequence analysis.
RNA Expression Analysis by Quantitative Real-Time PCR
Total RNA was extracted from paraffin-embedded cervical cancer samples from 39 HPV16-positive patients with sufficient tumor material present in the paraffin block (>70%). Samples were treated twice with 1 ml xylene for 10 min, washed with 100% ethanol and dried at 60 °C. Samples were digested at 80 °C for 30 min using 20 mM Tris, 1 mM EDTA and 2% SDS, and incubated overnight at 56 °C with 1 mg/ml RNAse-free proteinase K (Invitrogen). The next day, proteinase K was added again and the samples were incubated for 3–4 h. Subsequently, TRIzol (Invitrogen) and chloroform were added and RNA was isolated according to the manufacturer's protocol. Samples were treated with DNAse and reverse transcription was performed as described previously.28
Primers for HPV16 E5 and HPV16 E6 were designed according to Kivi et al29 and de Boer et al,28 respectively, and primers for EGFR were designed online (http://www.roche.com). All primer sequences are shown in the Supplementary information available at Modern Pathology's website. Each reaction was performed in duplicate on cDNA from HPV16-positive tumor specimens and included a standard curve of cDNA derived from five formalin-fixed, paraffin-embedded cervical cancer cell lines, namely CaSki, SiHa, CSCC1, C33 and HeLa. Quantitative PCR reactions were performed with an iCycler iQ detection system and analyzed using Optical Software (Biorad, Richmond, CA, USA). The expression of four reference genes (ACTB, HNRPM, RPL11 and UBC) was measured and the most stably expressed genes were identified with GeNorm.30 The geometric mean of ACTB and RPL11 was determined and expression data were normalized and log-transformed before statistical analysis.
Statistical Analysis
Statistical analyses were performed using the SPSS statistical program (Version 16.0 for Windows; SPSS Inc., Chicago, IL, USA). Significance tests were two-sided and statistical significance was assumed when P<0.05, corresponding to 95% confidence intervals (CI). The chi-squared (χ2) test was used for comparison of categorical data. Correlation analysis was performed using the Pearson's correlation test. Survival plots were made using the Kaplan–Meier method, testing significant differences using the log-rank test.
Results
Study Population
A total of 166 patients with cervical cancer were enrolled. Clinicopathological characteristics of patients are summarized in Table 1 . Median follow-up time was 113 months (range, 3–258) for all patients and 139 months (range, 3–258) for patients alive at the time of data collection.
Association between Clinicopathological Factors and Protein Expression of EGFR, mEGFR and pEGFR
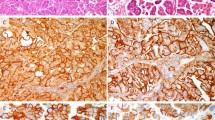
To quantify EGFR expression, immunohistochemical staining of EGFR was scored for EGFR, mEGFR and pEGFR. Representative examples of immunoreactivity are shown in Figure 1. Out of 166 patients, 14 patients showed normal epithelium next to the tumor. Of these 14 patients, the epithelium stained positive (normal expression, clearly distinguishable from the overexpression in the tumor) for nine patients and weak or negative for five patients. The staining was predominantly membranous. EGFR overexpression in cervical cancer specimens was defined as moderate or strong staining intensity. The distribution of EGFR, mEGFR and pEGFR staining is shown in Table 2 . pEGFR expression correlated with EGFR (P=0.008) and mEGFR (P=0.001) expression, and EGFR expression correlated with mEGFR expression (P<0.001). Both EGFR and mEGFR overexpression were observed more frequently in squamous cell carcinoma than in adeno(squamous) carcinoma (P=0.002 and <0.001, respectively), whereas pEGFR expression was not related to histopathological diagnosis (P=0.583, Table 2). EGFR overexpression was positively associated with tumor size greater than 40 mm (P=0.036) and tumor node metastasis (TNM) stage (P=0.005). mEGFR overexpression was positively associated with tumor size (P=0.021), lymph node positivity (P=0.004), vaso-invasion (P=0.043), parametrial invasion (P=0.009) and TNM stage (P=0.002). pEGFR expression was not associated with HPV type, tumor size, lymph node positivity, vaso-invasion, infiltration depth, parametrial invasion or TNM stage.
Immunohistochemical staining of epidermal growth factor receptor (EGFR) and phosphorylated EGFR (pEGFR) in squamous cell carcinoma of the cervix. Cytoplasmic/membrane staining of EGFR and membrane staining of EGFR (EGFR and mEGFR, respectively) were scored as strong (a), moderate (b), weak (c) or negative (d). pEGFR at Tyr1173 (pEGFR) was scored as positive (e) or negative (f). Original magnification × 400.
During the follow-up period, 45 patients died of cervical cancer (29%). The 5-year disease-specific survival rate was 71% in patients overexpressing EGFR compared with 83% in patients not overexpressing EGFR (P=0.084, Figure 2a), 73% in patients expressing pEGFR compared with 76% in pEGFR-negative patients (P=0.529, Figure 2b) and 72% in patients overexpressing mEGFR compared with 87% in patients not overexpressing mEGFR (P=0.027, Figure 2c).
Disease-specific survival in patients with epidermal growth factor receptor (EGFR) overexpression (a), phosphorylated EGFR (pEGFR) overexpression (b), membrane staining of the EGFR (mEGFR) overexpression (c) and mEGFR overexpression for human papillomavirus (HPV)16-positive patients (d), as well as high versus low HPV16 E5 (e) and HPV16 E6 (f) mRNA expression. High and low mRNA expression was based on the 75th percentile (p75). Sample sizes differ, depending on techniques used.
To analyze whether HPV positivity influences the association between EGFR expression and survival, we conducted survival analyses for the most prominent HPV types associated with cervical cancer (HPV16 and HPV18). For HPV16-positive patients (N=79), mEGFR overexpression was associated with disease-specific survival (P=0.029, Figure 2d), whereas EGFR and pEGFR were not. For HPV18-positive patients (N=31), EGFR, mEGFR and pEGFR expression were not associated with disease-specific survival (data not shown). Also, for patients with other HPV types (N=25) or HPV-negative patients (N=11), EGFR, mEGFR and pEGFR expression were not associated with disease-specific survival (data not shown).
Mechanism Leading to EGFR Overexpression
To analyze whether gene amplification is the predominant underlying mechanism for EGFR overexpression in cervical cancer, we performed FISH on a tissue array of 103 cervical cancer patients. In cervical cancer cells, EGFR was not amplified, when adjusted for ploidy of chromosome 7 (Figure 3). However, chromosome 7 aneuploidy was associated with mEGFR overexpression (P=0.013, Table 3 ).
A representative example of chromosome 7 aneuploidy with non-amplified epidermal growth factor receptor (EGFR). Fluorescence in situ hybridization on paraffin-embedded cervical cancer sample with a nucleus (arrow) containing four copies of chromosome 7 (red) and corresponding numbers of EGFR (green).
To assess whether mutations in EGFR were associated with EGFR overexpression, exon 18, 19, 20 and 21 were sequenced. For 96 patients, mutation analysis was performed on DNA isolated from flow-sorted, pure tumor cell subpopulations of paraffin-embedded cervical cancer specimens to ensure that contaminating normal cells do not interfere with the results. Although a common single-nucleotide polymorphism was apparent in exon 20 (rs1050171), analysis did not reveal any mutations in exons 18–21.
We also investigated the relation between HPV oncogenes and EGFR overexpression. As the association between disease-specific survival and mEGFR overexpression was strongest in HPV16-positive patients, mRNA expression levels of the HPV16 oncogenes E5 and E6 were determined. Furthermore, EGFR mRNA expression was measured to assess whether EGFR overexpression is associated with HPV16 E5 and E6 mRNA at the transcriptional level. mRNA expression levels of EGFR, HPV16 E5 and E6 were available for 32, 29 and 27 HPV16-positive patients, respectively. All values were corrected for the geometric mean of two reference genes, RPL11 and ACTB. Expression levels of HPV16 E5 and E6 were correlated (R2=0.528, P<0.001), but EGFR mRNA expression showed no correlation with E5 (R2=0.073, P=0.212) or E6 expression (R2=0.011, P=0.615). HPV16 E6 mRNA expression was positively associated with mEGFR protein expression (P=0.013, Table 3). High expression of HPV16 E5 and high expression of EGFR mRNA were not associated with EGFR protein expression (Table 3).
Association between Clinicopathological Factors and Chromosome 7 Aneuploidy, mRNA Expression of HPV16 E5, HPV16 E6 and EGFR
Chromosome 7 aneuploidy was positively associated with tumor size greater than 40 mm (P=0.006), but not with histopathology, TNM stage, infiltration depth, vaso-invasion, parametrial infiltration, lymph node positivity or prognosis (data not shown).
On the basis of the normal distribution of log-transformed expression data, an independent samples t-test was used to examine associations between E5, E6 and EGFR mRNA expression and clinicopathological factors. High HPV16 E6 expression was associated with vaso-invasion (mean difference=2.84 nSQ, 95% CI: 1.10–4.58, P=0.003) and infiltration depth of ≥15 mm (mean difference=3.00 nSQ, 95% CI: 1.04–4.96, P=0.004). HPV16 E6 expression was positively correlated with TNM stage (R2=0.172, P=0.039). No other associations were found for HPV16 E5, E6 and EGFR mRNA expression.
To establish whether high HPV16 oncogene expression was associated with survival, data were categorized according to mRNA expression greater and smaller than the 75th percentile. In contrast to EGFR mRNA expression (P=0.943), high expression of HPV16 E5 and E6 were associated with decreased disease-specific survival (P=0.045, Figure 2e and P=0.047, Figure 2f, respectively).
Discussion
We have investigated the association between EGFR expression and survival, as well as systematically analyzed the mechanisms responsible for EGFR overexpression in cervical cancer. Our data showed that mEGFR overexpression was associated with reduced survival in 166 cervical cancer patients. No mutations in EGFR were found. Overexpression of mEGFR was associated with aneuploidy of chromosome 7 and high expression of HPV16 E6 mRNA.
The proportion of cervical cancer patients overexpressing EGFR in this study is consistent with previous reports.4, 5, 6, 15 The association between mEGFR overexpression and prognosis has been reported previously with EGFR either as a single marker for decreased survival, or in combination with other biomarkers, such as COX-2.4, 15, 31 Lee et al32 compared cytoplasmic and membrane staining of EGFR and showed that only increased membrane staining was associated with impaired survival. Our results confirm that in cervical cancer only mEGFR overexpression and not pEGFR or cytoplasmic EGFR expression is negatively associated with prognosis.
EGFR is overexpressed in other cancer types, including glioblastoma multiforme, non-small-cell lung cancer and head and neck squamous cell cancer. In non-small-cell lung cancer, exons 18–21 are mutation hotspots, with deletions in exon 19 and the L858R mutation in exon 21 responsible for 85–90% of all EGFR mutations.13, 33 In head and neck squamous cell cancer, in which HPV infection contributes to ∼40% of the cases, EGFR mutations were found in 16% of all cases, but no EGFR mutations were detected in HPV-positive tumors.34, 35 These results suggest that the mechanism for EGFR overexpression in HPV-positive tumors is not associated with mutations in EGFR. In accordance with previous studies, we found no EGFR mutations in 96 cervical cancer specimens tested.17 As DNA was isolated from flow-sorted, pure tumor cells, we ensured that the probability of misinterpreting the results owing to dilution of tumor DNA with normal/stromal DNA was non-existent.
In glioblastoma multiforme, gene amplification is the predominant underlying mechanism of EGFR overexpression.36 Previously, Marzano et al18 reported that EGFR was not amplified in cervical cancer, when EGFR signals were corrected for chromosome 7 centromeric signals. However, EGFR protein expression was not analyzed in their study, so the effect of chromosome 7 aneuploidy could not be determined. We showed that although EGFR was not amplified in cervical cancer, chromosome 7 aneuploidy was associated with mEGFR overexpression.
Furthermore, our data also showed that HPV16 E6 expression was associated with mEGFR overexpression. Increased EGFR mRNA and protein levels have been reported in cervical cancer specimens, compared with normal cervical epithelium.6, 37 Immortalization of normal keratinocyte cell lines by transfection with HPV16 E6 showed upregulation of the EGFR, whereas HPV16 E5 and HPV16 E7 and HPV6 E6 did not influence EGFR expression.23, 38 Correspondingly, transfection of the cervical cancer cell line HeLa with antisense E6/E7 sequences resulted in the downregulation of the EGFR.39 As the E6 protein is known to target p53 for degradation, it has been suggested that HPV16 E6 regulates EGFR expression through p53.23 Indeed, inhibition of p53 was shown to increase EGFR expression in HPV16-immortalized human keratinocytes, supporting this hypothesis.40
To substantiate previous in vitro reports showing that the E5 protein increases EGFR expression by inhibiting the interaction of c-Cbl with the activated EGFR, thus decreasing EGFR degradation,22 we studied E5 expression in our patient cohort. Transgenic mouse models showed that expression of HPV16 E5 induced epithelial tumors and that this function of E5 was dependent on EGFR expression.41 However, although in our study high E5 and E6 mRNA expression were both associated with poor survival, we could not substantiate an association between HPV16 E5 and mEGFR expression. This could result from the fact that HPV16 E5 plays an important role in early carcinogenesis, or from the fact that E5 and E6 expression levels were strongly correlated.
In conclusion, we present a comprehensive study on the molecular mechanisms of EGFR overexpression in cervical cancer, using state-of-the-art techniques applied on archival, paraffin-embedded material of a large cohort of patients. High mEGFR protein expression as well as high HPV16 E5 and E6 mRNA levels were shown to predict an unfavorable prognosis in cervical cancer. Furthermore, our data showed for the first time that overexpression of mEGFR is influenced by chromosome 7 aneuploidy and HPV16 E6 mRNA expression.
References
Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108.
Eifel PJ, Winter K, Morris M, et al. Pelvic irradiation with concurrent chemotherapy versus pelvic and para-aortic irradiation for high-risk cervical cancer: an update of radiation therapy oncology group trial (RTOG) 90–01. J Clin Oncol 2004;22:872–880.
Green J, Kirwan J, Tierney J, et al. Concomitant chemotherapy and radiation therapy for cancer of the uterine cervix. Cochrane Database Syst Rev 2005;CD002225.
Kim GE, Kim YB, Cho NH, et al. Synchronous coexpression of epidermal growth factor receptor and cyclooxygenase-2 in carcinomas of the uterine cervix: a potential predictor of poor survival. Clin Cancer Res 2004;10:1366–1374.
Oh MJ, Choi JH, Kim IH, et al. Detection of epidermal growth factor receptor in the serum of patients with cervical carcinoma. Clin Cancer Res 2000;6:4760–4763.
Mathur SP, Mathur RS, Young RC . Cervical epidermal growth factor-receptor (EGF-R) and serum insulin-like growth factor II (IGF-II) levels are potential markers for cervical cancer. Am J Reprod Immunol 2000;44:222–230.
Ono M, Kuwano M . Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs. Clin Cancer Res 2006;12:7242–7251.
Cetuximab approved by FDA for treatment of head and neck squamous cell cancer. Cancer Biol Ther 2006;5:340–342.
Dolgin E . FDA narrows drug label usage. Nature 2009;460:1069.
Johnson JR, Cohen M, Sridhara R, et al. Approval summary for erlotinib for treatment of patients with locally advanced or metastatic non-small cell lung cancer after failure of at least one prior chemotherapy regimen. Clin Cancer Res 2005;11:6414–6421.
Cohen MH, Williams GA, Sridhara R, et al. United States food and drug administration drug approval summary: gefitinib (ZD1839; Iressa) tablets. Clin Cancer Res 2004;10:1212–1218.
Nicholas MK, Lukas RV, Jafri NF, et al. Epidermal growth factor receptor—mediated signal transduction in the development and therapy of gliomas. Clin Cancer Res 2006;12:7261–7270.
Rosell R, Taron M, Reguart N, et al. Epidermal growth factor receptor activation: how exon 19 and 21 mutations changed our understanding of the pathway. Clin Cancer Res 2006;12:7222–7231.
del Campo JM, Prat A, Gil-Moreno A, et al. Update on novel therapeutic agents for cervical cancer. Gynecol Oncol 2008;110:S72–S76.
Kersemaekers AM, Fleuren GJ, Kenter GG, et al. Oncogene alterations in carcinomas of the uterine cervix: overexpression of the epidermal growth factor receptor is associated with poor prognosis. Clin Cancer Res 1999;5:577–586.
Noordhuis MG, Eijsink JJ, Ten Hoor KA, et al. Expression of epidermal growth factor receptor (EGFR) and activated EGFR predict poor response to (chemo)radiation and survival in cervical cancer. Clin Cancer Res 2009;15:7389–7397.
Arias-Pulido H, Joste N, Chavez A, et al. Absence of epidermal growth factor receptor mutations in cervical cancer. Int J Gynecol Cancer 2008;18:749–754.
Marzano R, Corrado G, Merola R, et al. Analysis of chromosomes 3, 7, X and the EGFR gene in uterine cervical cancer progression. Eur J Cancer 2004;40:1624–1629.
Howie HL, Katzenellenbogen RA, Galloway DA . Papillomavirus E6 proteins. Virology 2009;384:324–334.
McLaughlin-Drubin ME, Münger K . The human papillomavirus E7 oncoprotein. Virology 2009;384:335–344.
Zyzak LL, MacDonald LM, Batova A, et al. Increased levels and constitutive tyrosine phosphorylation of the epidermal growth factor receptor contribute to autonomous growth of human papillomavirus type 16 immortalized human keratinocytes. Cell Growth Differ 1994;5:537–547.
Zhang B, Srirangam A, Potter DA, et al. HPV16 E5 protein disrupts the c-Cbl–EGFR interaction and EGFR ubiquitination in human foreskin keratinocytes. Oncogene 2005;24:2585–2588.
Akerman GS, Tolleson WH, Brown KL, et al. Human papillomavirus type 16 E6 and E7 cooperate to increase epidermal growth factor receptor (EGFR) mRNA levels, overcoming mechanisms by which excessive EGFR signaling shortens the life span of normal human keratinocytes. Cancer Res 2001;61:3837–3843.
Koopman LA, Szuhai K, van Eendenburg J, et al. Recurrent integration of human papillomaviruses 16, 45 67 near translocation breakpoints in new cervical cancer cell lines. Cancer Res 1999;59:5615–5624.
Haralambieva E, Kleiverda K, Mason DY, et al. Detection of three common translocation breakpoints in non-Hodgkin's lymphomas by fluorescence in situ hybridization on routine paraffin-embedded tissue sections. J Pathol 2002;198:163–170.
Waye JS, England SB, Willard HF . Genomic organization of alpha satellite DNA on human chromosome 7: evidence for two distinct alphoid domains on a single chromosome. Mol Cell Biol 1987;7:349–356.
Corver WE, Ter Haar NT, Dreef EJ, et al. High-resolution multi-parameter DNA flow cytometry enables detection of tumour and stromal cell subpopulations in paraffin-embedded tissues. J Pathol 2005;206:233–241.
de Boer MA, Jordanova ES, Kenter GG, et al. High human papillomavirus oncogene mRNA expression and not viral DNA load is associated with poor prognosis in cervical cancer patients. Clin Cancer Res 2007;13:132–138.
Kivi N, Greco D, Auvinen P, et al. Genes involved in cell adhesion, cell motility and mitogenic signaling are altered due to HPV 16 E5 protein expression. Oncogene 2008;27:2532–2541.
Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034 (online article).
Hale RJ, Buckley CH, Gullick WJ, et al. Prognostic value of epidermal growth factor receptor expression in cervical carcinoma. J Clin Pathol 1993;46:149–153.
Lee CM, Lee RJ, Hammond E, et al. Expression of HER2neu (c-erbB-2) and epidermal growth factor receptor in cervical cancer: prognostic correlation with clinical characteristics, and comparison of manual and automated imaging analysis. Gynecol Oncol 2004;93:209–214.
Riely GJ, Politi KA, Miller VA, et al. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res 2006;12:7232–7241.
Kong CS, Narasimhan B, Cao H, et al. The relationship between human papillomavirus status and other molecular prognostic markers in head and neck squamous cell carcinomas. Int J Radiat Oncol Biol Phys 2009;74:553–561.
Na II, Kang HJ, Cho SY, et al. EGFR mutations and human papillomavirus in squamous cell carcinoma of tongue and tonsil. Eur J Cancer 2007;43:520–526.
Kuan CT, Wikstrand CJ, Bigner DD . EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr Relat Cancer 2001;8:83–96.
Manavi M, Hudelist G, Fink-Retter A, et al. Gene profiling in Pap-cell smears of high-risk human papillomavirus-positive squamous cervical carcinoma. Gynecol Oncol 2007;105:418–426.
Wilding J, Vousden KH, Soutter WP, et al. E-cadherin transfection down-regulates the epidermal growth factor receptor and reverses the invasive phenotype of human papilloma virus-transfected keratinocytes. Cancer Res 1996;56:5285–5292.
Hu G, Liu W, Mendelsohn J, et al. Expression of epidermal growth factor receptor and human papillomavirus E6/E7 proteins in cervical carcinoma cells. J Natl Cancer Inst 1997;89:1271–1276.
Bheda A, Creek KE, Pirisi L . Loss of p53 induces epidermal growth factor receptor promoter activity in normal human keratinocytes. Oncogene 2008;27:4315–4323.
Genther Williams SM, Disbrow GL, Schlegel R, et al. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Res 2005;65:6534–6542.
Acknowledgements
We gratefully acknowledge Jan Molenaar for his help with the collection of follow-up data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Schrevel, M., Gorter, A., Kolkman-Uljee, S. et al. Molecular mechanisms of epidermal growth factor receptor overexpression in patients with cervical cancer. Mod Pathol 24, 720–728 (2011). https://doi.org/10.1038/modpathol.2010.239
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2010.239