Abstract
Background
Cervical carcinogenesis is mediated by the HPV-E6 and E7 oncoproteins, considered as biomarkers usable in managing screen-positive women.
Methods
We conducted a systematic review and meta-analysis assessing the accuracy of HPV-E6/E7-oncoprotein tests to detect underlying cervical-precancer and cancer. We included studies reporting data on oncoprotein test accuracy detecting cervical intraepithelial neoplasia grade 3 or worse. Random effects logistic regression models were applied for pooling absolute and relative accuracy.
Results
Twenty-two studies were included. Sensitivity and specificity estimates ranged from 54.2% (95%CI: 45.2–63.0) to 69.5% (95%CI:60.8–76.9) and from 82.8% (95%CI: 50.4–95.8) to 99.1 (95%CI: 98.8–99.3), respectively in the population irrespective of HPV status. Higher sensitivity estimates ranging from 60.8% (95%CI: 49.6–70.9) to 75.5% (95%CI: 71.7–78.9) but lower specificity estimates ranging from 83.7% (95%CI: 76.1–89.3) to 92.1% (95%CI: 88.5–94.6) were observed in studies enrolling high-risk-HPV-positive women. Studies recruiting only HIV-positive women showed a pooled sensitivity of 46.9% (95%CI: 30.6–63.9) with a specificity of 98.0% (95%CI: 96.8–98.7).
Conclusions
The high specificity of oncoprotein tests supports its use for triaging HPV-positive women. However, oncoprotein-negative women would not be recommended to undertake routine screening, requiring further follow-up. Large-scale and longitudinal studies are needed to further investigate the role of E6/E7-oncoprotein detection in predicting the risk of developing cervical pre-cancer and cancer.

Similar content being viewed by others
Introduction
Cervical cancer is preventable, yet more than 600,000 new cases are diagnosed and approximately 350,000 deaths from the disease occur each year [1]. Cervical cancer is ranked as the fourth most common cancer worldwide and is, therefore, a major public health concern [2]. Persistent infection with carcinogenic Human Papilloma Virus (HPV) types is a necessary cause of cervical cancer [3]. HPV infection can lead to precancerous lesions such as cervical intraepithelial neoplasia 2, or 3 (CIN2, CIN3) which, if left untreated, can eventually progress to cervical cancer. Many women infected with HPV will naturally clear the infection within a year through their immune system, but in considerable proportion, the infection will persist for more than 2 years possibly leading to precancerous lesions and eventually transforming into cancer [4,5,6,7].
Most of the available HPV screening tests cannot differentiate between transient and persistent HPV infection and this is important for building an optimal and clinically relevant screening and follow-up approach. The 2021 guidelines from the World Health Organization (WHO) recommend using a high performance test for primary cervical cancer screening, such as HPV DNA assays [8]. However, due to the high prevalence of transient infections, HPV testing will increase the number of referrals to colposcopy and will generate unnecessary anxiety for women who are not at high risk of developing cervical cancer, highlighting the need for a second test to triage HPV positive women [9, 10]. Given the high global HPV prevalence, and especially in high-risk groups such as women living with HIV (WLWH) or in women from a low-and-middle-income country (LMIC), an accurate test is urgently needed to identify and triage those women who are at high risk of developing true disease. Current screening programmes in LMICs have limited resources and follow-up after the positivity of the first screening test can be challenging.
It is now well established that the viral oncoproteins E6 and E7 are necessary to promote and maintain cervical carcinogenesis and are both overexpressed during cervical transformation. Indeed, they have a synergic action and can activate many cancer hallmarks [11, 12]. The oncoprotein E6 has a major role in cell immortalization. It can bind to the tumour suppressor protein p53, which is usually activated in response to DNA damages in normal cells. This binding degrades the protein p53, which prevents the cell from undergoing apoptosis [13]. The oncoprotein E7 also interacts with numerous host proteins, and more specifically, it can bind to the cellular tumour suppressor, pRb [14, 15]. This binding to pRb leads to its degradation which prevents it from inactivating transcription factors and therefore favouring cell proliferation [16]. The overexpression of E6 and E7 leading to the accumulation of genetic errors in the cells over time in combination with viral gene deregulation expression, drives to the invasive cancer phenotype [12, 17, 18]. Oncoproteins E6 and E7 are, therefore, interesting candidates as markers of persistent HPV infection and associated pre-cancerous lesions. It is important to note that these oncoproteins E6 and E7 differ by HPV type and, therefore, tests must target oncoproteins from diverse oncogenic types to assure sufficient sensitivity. The clinical accuracy of E6 or E7 DNA and RNA assays have been evaluated in several comprehensive meta-analyses but the performance of tests targeting the E6 and E7 proteins has not been systematically reviewed yet [9, 19].
We aimed to synthesize the available and most up-to-date evidence on the accuracy of tests targeting HPV E6/E7 oncoproteins for CIN3+ (CIN2+) detection through a systematic review of the literature and meta-analysis.
Methods
Search strategy and selection criteria
The current systematic review includes a meta-analysis and was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) requirements [20, 21]. The Population, Intervention, Comparator and Outcome (PICO) components were used to define the eligibility criteria (Supplementary Materials, page 3). Cohorts, case-control, cross-sectional diagnostic accuracy studies, and clinical trials were considered. Studies were included if they were reporting original data; if the disease of interest was cervical precancer (defined as cervical intraepithelial neoplasia of grade 2 or 3 or worse (CIN2 + /CIN3 + )); if they used a test targeting the E6 and/or the E7 oncoprotein; and if the gold standard for diagnosis was colposcopy guided histology or negative colposcopy. Articles were excluded if studies were in-vitro, animal-based or molecular; or if they assessed a proof of concept for the oncoprotein test development. Samples different from exfoliated cervical/vaginal cells such as sera or tissue samples were not considered.
A multi-database search was conducted, looking into PubMed, Embase and Web of Science for eligible publications in all languages without limitation of publication date until December 2021. The following search terms were used and combined: cervical cancer AND oncoproteins AND HPV AND screening. The Mesh function [MeSH] and Title and Abstract [tiab] were applied to refine the search (Supplementary Materials, pages 4–11). Reference lists of included articles and reviews on the topic were also manually screened. The grey literature was searched such as conference abstracts, thesis dissertations, reports, and local institution libraries to assess potential publication bias. More specifically, the online IARC library, EThOS (UK E-Theses Online Service), and the Open Grey website were thoroughly searched. Extensive searches were conducted to retrieve the full text of eligible articles, however, in case of unavailability, there were not included. In the case of multiple reports from the same study, the most recent one with the longest follow-up period or the one reporting the most comprehensive data was included. If unpublished data, researchers were systematically contacted to obtain as much information as possible. Summary estimates were sought for each included study. Titles and abstracts identified through the 3 electronic databases were screened by one reviewer (LD) and any questions or doubts were solved by discussion with a second reviewer (MLR).
Data analysis
Titles and abstracts were screened against the eligibility criteria and for all eligible articles, full texts were obtained when possible. They were then independently examined in detail by two reviewers (LD and IJ) for their eligibility in the review. All papers excluded at this second stage of the selection process were documented along with the reasons for exclusion. Any questions and doubts were resolved through discussion or the intervention of a third reviewer (M. Arbyn). Moreover, studies requiring more calculation for data extraction were reviewed by a third reviewer (MLR). All identified references were stored into EndNote bibliographic software for further assessment and handling. Duplicate citations were removed manually. Hand-searching was preferred to the automatic de-duplicating option from EndNote as this could lead to removing articles that should not be deleted [22]. References were then transferred to Rayyan, an online application that allowed facilitating the screening process between reviewers [23]. For each included study, data was extracted by two independent reviewers (LD and IJ) and reported in a shared table (Supplementary materials, page 12. If any conflicts arose, they were resolved by discussion and submitted to M. Arbyn for final judgment, if necessary. Efforts were made to minimize reporting bias (Supplementary Materials, page 12).
The QUADAS-2 checklist was used to assess the quality of included diagnostic studies [24] (Supplementary Materials, pages 30–31).
Random effect logistic regression models were applied for pooling of sensitivity and specificity estimates and accounting for the intrinsic correlation between the log odds of the true and false positivity rate [25]. Results were displayed through forest plots. Separate meta-analyses were conducted for the outcomes CIN3+ (primary) and CIN2+. Analyses were stratified by the type of recruited study population, that is, screening, triage, colposcopy referral or convenience sample. If the results from the stratified analysis suggested some differences, a meta-regression was performed to formally explore differences with the covariate of interest. Heterogeneity statistic I2 was used to measure heterogeneity among studies. Studies targeting two different sources of populations—such as women coming from (i) a screening approach and (ii) women referred to colposcopy—were included in the convenience sample group. Of note, we invite the reader to interpret sub diamond results with caution when heterogeneity is high given the different design methodology of the included studies. Additional analyses were performed assessing some covariates as potential explanatory variables of accuracy: study population (population irrespective of HPV status, triage of hrHPV-positive or HPV16/18-positive), QUADAS quality items, countries’ incomes, and sample storage temperature. A separate analysis was conducted for studies including only women living with HIV (WLWH).
Relative estimates were also displayed through forest plots, comparing the performance of oncoprotein testing to that of comparator tests whenever reported, and we explicitly mentioned the comparator test in corresponding figures. These comparator tests included HPV DNA/RNA based tests; visual inspection tests or molecular-based tests.
If studies reported accuracy estimates separately for different populations such as (i) in the population irrespective of HPV status (ii) in hrHPV-positive women, or (iii) in HPV16/18-positive women; they were considered as three different studies in the analysis. Similarly, if studies reported accuracy estimates for different specimen types (clinician vs self-collected samples, stored in PreservCyt vs SurePath), data from clinician-collected samples in PreservCyt were extracted as the majority of included studies used this approach. When multiple HPV tests were used, we extracted the data for those HPV assays that are clinically validated [19]. Wherever HC2 (QIAGEN, Germantown, United States) and CareHPV (QIAGEN, Hilden, Germany) testing were used, HC2 testing was preferred as it has better accuracy than CareHPV for CIN3+ (CIN2+) detection [9]. Similarly, when cervical samples were collected with different devices, dry swab was the preferred collection approach rather than brush in liquid-based preservative medium. Finally, if a study reported accuracy estimates for two or more different oncoprotein tests, they were reported separately as two studies in the analysis.
All data were analysed using StataSE17 software.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
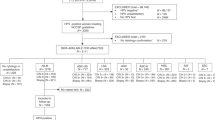
The search identified 6063 records which were eligible for title and abstract screening. Once duplicates were removed, 3861 records were screened from which 47 were screened in full text. Finally, 22 of those met the inclusion criteria and were therefore considered in the review for final analysis (Fig. 1) [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47]. These studies were conducted between 1999 and 2020, and published between 2010 and 2020 (median 2017, IQR 2016–2019). Considering CIN2+ and CIN3+ endpoints together, six studies reported the accuracy of the oncoprotein test within a screening population, seven within a colposcopy referral population, and nine within a convenience sample (Supplementary Materials, pages 13–15). Two studies targeted only WLWH. In ten studies, the accuracy of the test was assessed in hrHPV+ women and six studies restricted the analysis to specifically HPV16/18+ women. Characteristics of the included studies are reported in Table 1 and all technical principles of oncoprotein tests are detailed in Supplementary Materials Pages 16–17.
Among the 11 studies reporting the oncoprotein test accuracy for CIN3+ in the population irrespective of HPV status, the meta-analysis revealed sensitivity and specificity estimates of 54.2% (95%CI: 45.2–63.0) and 99·1% (95%CI: 98.8–99.3) for screening populations compared to 61.7% (95%CI: 52.4–70.2) and 82.8% (95%CI: 50.4–95.8) for colposcopy referral populations and finally 69.5% (95%CI: 60.8–76.9) and 94.4% (95%CI: 78.7–98.7) for convenience samples (Fig. 2a). These overall estimates did not significantly differ when stratified by population type as shown by the covariate analysis in the Supplementary Materials Page 18.
In hrHPV-positive women, the sensitivity was lower in colposcopy referral populations (60.8%) than in screening and convenience samples (74.6% and 75.5%, respectively), with specificities ranging from 83.7% to 92.1% across the different populations (Fig. 2b). However, these accuracy differences across the populations were not statistically significant (Supplementary Materials Page 19).
When restricted to HPV16/18+ women (Fig. 2c), the sensitivity was similar in the colposcopy referral group (60.8%, 95%CI: 50.8–70.0) and higher in the convenience sample (81%, 95%CI: 63.1–91.4) while the specificity was lower (78.4%, 95%CI: 74.6–81.8; 82.1%, 95%CI: 69.9–90.1, respectively) than in the population irrespective of HPV status and hrHPV-positive women. The covariate analysis did not show any significant accuracy differences between the two population groups (Supplementary Materials Page 20). Regarding the two studies including only WLWH, the pooled sensitivity estimate was 46.9% (95%CI: 30.6–63.9) with a corresponding specificity of 98.9% (95%CI: 96.8–98.7) (Fig. 2d).
In all populations, similar sensitivities were obtained with different sample storage temperatures, but the specificity increased from 68% with a storage at 4 °C to 98% with a storage at −60–80 °C as displayed in the Supplementary Materials Pages 21–23 (The meta-regression did not show significant differences in accuracy across the different groups of storage).
Most of the studies were conducted in upper-middle income countries (64%) with few in low-middle and upper-middle income countries (18%) as well as in (Upper-middle) high-income countries (18%). Stratification by income level did not explain study heterogeneity (Supplementary Materials, Page 24).
Eight studies were retrieved in which accuracy of the oncoprotein test to detect CIN3+ could be compared to that of other screening tests in the population irrespective of HPV status (Fig. 3a). When compared to hrHPV DNA, HPV16/18 DNA testing, mRNA testing, and cytology, oncoprotein testing was significantly less sensitive but more specific: relative sensitivity estimates of 0.78 (95%CI: 0.73–0.83); 0.71 (95%CI: 0.62–0.80); 0.58 (95%CI: 0.48–0.68); 0.73 (95%CI: 0.66–0.81), respectively, and relative specificity estimates of 1.34 (95%CI: 1.08–1.67); 1.78 (95%CI: 1.23–2.58); 1.66 (95%CI: 1.40–1.97); and 1.30 (95%CI: 1.06–1.59), respectively. Oncoprotein testing was 19% more sensitive and 6% more specific than visual inspection with acetic acid (VIA) for CIN3+ detection (relative sensitivity: 1.19, 95%CI: 0.92–1.55; relative specificity: 1.06, 95%CI: 1.06–1.07, p = 0.02).
a Population irrespective of HPV status; b hrHPV-positive women. Results from a meta-regression using as a covariate the type of screening test. ASCH+ Atypical Squamous Cells where High-grade squamous intra-epithelial lesions cannot be excluded, ASCUS+ Atypical Squamous cells of Undetermined Significance, HC2 Hybrid Capture 2, HSIL High grade Squamous Intraepithelial Lesion, RT Real Time.
The relative analysis from the six studies including hrHPV+ women showed similar results but were less marked (Fig. 3b). Oncoprotein testing was slightly less sensitive but more specific than cytology, genotyping (HPV DNA partial testing with LiPA 16/18 or Cobas HPV16/18; or extended genotyping testing with LiPA 8-types) and colposcopy with pooled relative sensitivity estimates of 0.89 (95%CI: 0.80–0.99), 0.86 (95%CI: 0.77–0.98), and 0.80 (95%CI: 0.59–1.07), respectively; and relative specificity of 1.28 (95%CI: 1.19–1.38), 1.22 (95%CI: 1.14–1.29), and 1.14 (95%CI: 1.04–1.26), respectively. In one study, oncoprotein testing was less sensitive than dual staining and HPV DNA testing (0.63, 95%CI: 0.40–0.97; 0.60, 95%CI: 0.48–0.72, respectively) but more specific (1.05, 95%CI: 0.99–1.12; 2.33, 95%CI: 2.16–2.53, respectively). Finally, oncoprotein testing showed a slightly better accuracy than VIA in one study with a relative sensitivity of 1.02 (95%CI: 0.78–1.33) and a relative specificity of 1.10 (95%CI: 1.07–1.14).
Results for the CIN2+ endpoint were similar to those of CIN3+ with the exception of oncoprotein testing that was found to be less sensitive than VIA in hrHPV-positive women (relative sensitivity: 0.52, 95%CI: 0.44–0.60, Supplementary Materials, Pages 25–29).
Quality assessment using the QUADAS tool
Overall, there was a wide spectrum of patient selection approaches across the included studies (55%), reflecting the populations described from different sources, that is, women mainly enrolled from a clinical setting, gynaecological department, referred after a screen-positive results or needing colposcopy and therefore not derived from a primary screening population (Supplementary Materials, Pages 30–31).
We found an increased sensitivity with increased risk of bias in patient selection for CIN3+ detection (54%, 58%, and 68% in studies with low, moderate, and high risk of bias, respectively) (Supplementary Materials, Page 32).
Significantly increased risk of screening test bias with increased reported sensitivity for CIN3+ was found (58% in low, 69% in moderate, and 73% in high risk of bias, Supplementary Materials, Page 33).
Finally, there was an increased sensitivity with increased risk of bias in reference testing with overall 62% sensitivity in studies with a low-risk score and 72% sensitivity in studies with a moderate risk score (Supplementary Materials, Page 34).
Discussion
The meta-analysis revealed a high specificity of oncoprotein testing, over 82% in the population irrespective of HPV status, in hrHPV-positive, in HPV16/18-positive and in WLWH. Moderate sensitivity estimates improved when the study population was restricted to hrHPV or HPV16/18-positive women. All estimates varied largely across studies with both sensitivity and specificity ranging from around 46.9% to 99.1%.
This wide observed variation in accuracy could be attributed to different factors such as type of oncoprotein test (targeting the E6 or E7 or both proteins, commercial or in-house developed test), sample storage and collection device. Accuracy estimates were improved when samples were stored between −60 °C and −80 °C. Sample collection device also varied between studies using dry Dacron swab or Cervex brush with a liquid-based medium but the lack of such data within the available studies did not allow further analyses considering these parameters. Studies from Kong et al., Schweizer et al., Wu et al., Yang et al., and Ara et al., reported sensitivities above 67% (range: 67.4% to 81.8%) [26, 32, 37, 43, 44]. A plausible reason for these high estimates could be explained by the study population. Indeed, most of these used a convenience sample of hrHPV-positive women including HPV16/18-positive women in the Kong study and enriched their population with cervical cancer cases in Schweizer, Wu, and Yang studies. Finally, women referred to colposcopy after an abnormal VIA result were included in one study [26]. Therefore, women included in these studies had higher probability of having high-grade lesions and thus were more prone to have elevated expression of oncoproteins with subsequent better detection from the oncoprotein testing. This is underpinned by the QUADAS analysis findings revealing high risk of bias in patient selection for Ara, Yang, Schweizer and Kong studies. It is worth noting that specificity in HPV16/18-positive women was lower than in the population irrespective of HPV status or in hrHPV-positive women (75% vs 94.2% vs 90.8%). This finding is consistent with the study from Rossi et al., showing that specificity decreases with high HPV prevalence. The results from the meta-regression that we conducted here showed that the slope is decreasing (−0.34, 95%CI: −1.69 -1.00) (Supplementary Materials Pages 35–36). Most of the included studies used the OncoE6 test, which is correlated to HPV types 16 and 18. OncoE6 testing could be used to triage HPV16/18-positive women perhaps to prioritise women positive for both HPD DNA and oncoprotein test compared to oncoprotein-negative women.
Our results showed that oncoprotein testing was significantly more specific than HPV DNA and mRNA testing (relative specificity of 1.34 and 1.66, respectively). This finding is consistent with results from several studies reporting stronger correlations between E6 oncoprotein expression and severity of the cervical lesion than the correlation between HPV positivity and lesion severity [37, 38, 48]. Wu et al. reported a positive association between HPV DNA viral load and expression of E6 oncoprotein in (pre-)cancer lesions as well as consistency regarding oncoprotein E6 positivity and hrHPV viral load and increased disease severity [43]. These findings highlight the potential of using oncoprotein detection as a marker of disease severity and progression towards CIN3 or cancer.
It has been shown that there is an increased number of cells with integrated HPV genome in cells progressing to malignancy [49]. A study from the Cancer Genome Atlas showed that HPV was integrated in all HPV18-positive cases and in 76% of HPV16-positive cases [50]. From our meta-analysis results, oncoprotein testing appears to have a low sensitivity compared to HPV DNA testing. The main hypothesis relies on the viral HPV DNA, which would have not been integrated within the host genome in precancerous lesions not progressing to cervical cancer. Therefore, the oncoproteins E6/E7 are not expressed -or are undetectable- and the oncoprotein test cannot detect them as the oncoproteins are only highly expressed in true pre-cancerous cells. Although it has been shown that integration is associated with progression, the integration process does not mean that episomal HPV disappears and, therefore, totally explains drop in the sensitivity of HPV testing. This is only a hypothesis and will require further studies to investigate. In some studies, false positivity of the oncoprotein test was high, possibly explained by technical limitations of the tests themselves as well as sampling methods, the number and quality of collected cells and the storage process [29, 32, 44]. However, we are aware that the comparison in sensitivity between oncoproteins and HPV DNA testing has not a strong rationale given that HPV infection is a necessary condition for the disease. The additional interest here includes triaging WLWH or immunocompromised HPV-positive women in general and especially in screen and treat schemes to avoid unnecessary treatment. Indeed, WLWH come back to care more regularly than HIV-negative women, thus the opportunity to be reassessed more often.
To date, the gold standard test for cervical cancer diagnosis is histological examination which provides clinical information such as stage of the disease. However, whether the cervical lesion will be more likely to progress, or regress, is still complex to predict. Hence, the detection of E6/E7 oncoproteins could add another level of information regarding the potential progress to carcinogenic phenotype and help identify women at higher risk of developing true disease.
Most of the studies were conducted in upper-middle income countries with a few in low-middle income countries or high-income countries. The test accuracy did not appear to be compromised by the income country level, revealing the possibility of performing the test in such resource limited settings.
This meta-analysis is the first reporting on the accuracy of oncoprotein testing. Data were extracted and revised by different reviewers and a rigorous QUADAS analysis was conducted. The literature search was neither restricted to language nor to publication date.
It is important to note that given the recruitment strategy, all included studies reporting accuracy in hrHPV-positive women could not be assessed as a triage setting. Indeed, not all included women were hrHPV-positive, and all did not have one or more triage test(s) as different diagnostic algorithms were applied. Similarly, studies including a convenience sample could not be assessed as a screening setting given that most of included women were an arbitrary case-mix derived from primary screening and from a clinical setting and referred from screen-positive results. Our review was restricted to cross-sectional outcomes that is, sensitivity and specificity of the oncoprotein test against the reference test with no available longitudinal data to evaluate. Finally, there was too much variation in reporting age, which precluded us to assess it as covariate of oncoprotein accuracy.
More research is needed to assess whether oncoprotein testing can distinguish between HPV driven lesions with higher potential of carcinogenesis and lesions with poor progression potential. The study from Zhang et al. reported on the role of the OncoE6 testing as a triage method for HPV positive women, to predict the risk of developing CIN3+. It showed that women with persistent hrHPV infection had 40 times the odds (95%CI = 11.8–135.5) of expressing the E6 oncoprotein compared to women with incident HPV infection during the 15-year follow-up and that there was an increased risk of subsequent HPV DNA persistence in women who tested positive for OncoE6 compared to women who tested negative (adjusted OR = 21.2, 95%CI = 5.2–86.4). Finally, more research focusing on the regressions rates of oncoprotein negative CIN3 lesions would help understanding of the longitudinal safety of oncoprotein testing.
In conclusion, this meta-analysis reveals a high specificity of oncoprotein testing across all groups (population irrespective of HPV status, hrHPV-positive, HPV16/18-positive, and WLWH), with moderate sensitivity estimates. The corresponding sensitivity estimates were comparable or higher to those of cytology and VIA for CIN3+ detection in primary screening. Oncoprotein testing may be useful for triaging HPV-positive women. However, the role of oncoprotein test should be further explored in populations such as WLWH for whom high specific screening and triage tests may be needed.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Singh D, Vignat J, Lorenzoni V, Eslahi M, Ginsburg O, Lauby-Secretan B, et al. Global estimates of incidence and mortality of cervical cancer in 2020: a baseline analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Glob Health. 2023;11:e197–e206.
Cancer Today GCO. Estimated number of new cases in 2020, worldwide, females, all ages 2020 [Available from: https://gco.iarc.fr/today/online-analysis-table?v=2020&mode=cancer&mode_population=continents&population=900&populations=900&key=asr&sex=2&cancer=39&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&include_nmsc=0&include_nmsc_other=1. Access Date: March 3, 2022.
Okunade KS. Human papillomavirus and cervical cancer. J Obstet Gynaecol. 2020;40:602–8.
Rodríguez AC, Schiffman M, Herrero R, Wacholder S, Hildesheim A, Castle PE, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst. 2008;100:513–7.
Kitchener HC, Almonte M, Gilham C, Dowie R, Stoykova B, Sargent A, et al. ARTISTIC: a randomised trial of human papillomavirus (HPV) testing in primary cervical screening. Health Technol Assess. 2009;13:1–150.
Plummer M, Schiffman M, Castle PE, Maucort-Boulch D, Wheeler CM. A 2-year prospective study of human papillomavirus persistence among women with a cytological diagnosis of atypical squamous cells of undetermined significance or low-grade squamous intraepithelial lesion. J Infect Dis. 2007;195:1582–9.
Rodríguez AC, Schiffman M, Herrero R, Hildesheim A, Bratti C, Sherman ME, et al. Longitudinal study of human papillomavirus persistence and cervical intraepithelial neoplasia grade 2/3: critical role of duration of infection. J Natl Cancer Inst. 2010;102:315–24.
World Health Organization. New recommendations for screening and treatment to prevent cervical cancer 2021 [Available from: https://www.who.int/news/item/06-07-2021-new-recommendations-for-screening-and-treatment-to-prevent-cervical-cancer. Access Date: March 3, 2022.
Arbyn M, Snijders PJ, Meijer CJ, Berkhof J, Cuschieri K, Kocjan BJ, et al. Which high-risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect. 2015;21:817–26.
Arbyn M, Ronco G, Anttila A M, Meijer CJ, Poljak M, Ogilvie G, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30:F88–F99.
Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46.
Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–82.
Araldi RP, Sant’Ana TA, Módolo DG, de Melo TC, Spadacci-Morena DD, de Cassia Stocco R, et al. The human papillomavirus (HPV)-related cancer biology: an overview. Biomed Pharmacother. 2018;106:1537–56.
Boccardo E, Baldi CVM, Carvalho AF, Rabachini T, Torres C, Barreta LA, et al. Expression of human papillomavirus type 16 E7 oncoprotein alters keratinocytes expression profile in response to tumor necrosis factor-alpha. Carcinogenesis. 2010;31:521–31.
Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–60.
Giarrè M, Caldeira S, Malanchi I, Ciccolini F, Leão MJ, Tommasino M. Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a-imposed G1 cell cycle Arrest. J Virol. 2001;75:4705–12.
Yeo-Teh NSL, Ito Y, Jha S. High-risk human papillomaviral oncogenes E6 and E7 target key cellular pathways to achieve oncogenesis. Int J Mol Sci. 2018;19:1706.
McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017;13:e1006211.
Arbyn M, Simon M, Peeters E, Xu L, Meijer C, Berkhof J, et al. 2020 list of human papillomavirus assays suitable for primary cervical cancer screening. Clin Microbiol Infect. 2021;27:1083–95.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. Bmj. 2009;339:b2700.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Bmj. 2021;372:n71.
Kwon Y, Lemieux M, McTavish J, Wathen N. Identifying and removing duplicate records from systematic review searches. J Med Libr Assoc. 2015;103:184–8.
Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan-a web and mobile app for systematic reviews. Syst Rev. 2016;5:210.
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36.
Nyaga VN, Arbyn M. Metadta: a Stata command for meta-analysis and meta-regression of diagnostic test accuracy data—a tutorial. Arch Public Health. 2022;80:95.
Ara R, Khatun S, Pervin S, Jahan M, Shahera U, Ferdous J, et al. Role of molecular biomarker human papilloma virus (HPV) E6 oncoprotein in cervical cancer screening. Gynecol Oncol. 2020;158:590–6.
Agorastos T, Chatzistamatiou K, Moysiadis T, Kaufmann AM, Skenderi A, Lekka I, et al. Human papillomavirus E7 protein detection as a method of triage to colposcopy of HPV positive women, in comparison to genotyping and cytology. Final results of the PIPAVIR study. Int J Cancer. 2017;141:519–30.
Chibwesha CJ, Frett B, Katundu K, Bateman AC, Shibemba A, Kapambwe S, et al. Clinical performance validation of 4 point-of-care cervical cancer screening tests in HIV-infected women in Zambia. J Low Genit Trac Dis. 2016;20:218–23.
Cuzick J, Ahmad AS, Austin J, Cadman L, Ho L, Terry G, et al. A comparison of different human papillomavirus tests in PreservCyt versus SurePath in a referral population-PREDICTORS 4. J Clin Virol. 2016;82:145–51.
de Oliveira CM, Musselwhite LW, Pantano ND, Vazquez FL, Smith JS, Schweizer J, et al. Detection of HPV E6 oncoprotein from urine via a novel immunochromatographic assay. Plos One. 2020;15:e0232105.
Ferrera A, Valladares W, Cabrera Y, de la Luz Hernandez M, Darragh T, Baena A, et al. Performance of an HPV 16/18 E6 oncoprotein test for detection of cervical precancer and cancer. Int J Cancer. 2019;145:2042–50.
Kong L, Xiao X, Lou H, Liu P, Song S, Liu M, et al. Analysis of the role of the human papillomavirus 16/18 E7 protein assay in screening for cervical intraepithelial neoplasia: a case control study. BMC Cancer. 2020;20:999.
Mariano VS, Lorenzi AT, Scapulatempo C, Stein MD, Resende JCP, Antoniazzi M, et al. A low-cost HPV immunochromatographic assay to detect high-grade cervical intraepithelial neoplasia. Plos One. 2016;11:e0164892.
Ndizeye Z, Menon S, Van Geertruyden J-P, Sauvaget C, Jacquemyn Y, Bogers J-P, et al. Performance of OncoE6 (TM) Cervical Test in detecting cervical precancer lesions in HIV-positive women attending an HIV clinic in Bujumbura, Burundi: a cross-sectional study. Bmj Open. 2019;9:e029088.
Qiao YL, Jeronimo J, Zhao FH, Schweizer J, Chen W, Valdez M, et al. Lower cost strategies for triage of human papillomavirus DNA-positive women. Int J Cancer. 2014;134:2891–901.
Rezhake R, Hu SY, Zhao S, Xu XQ, Zhao XL, Zhang L, et al. Eight-type human papillomavirus E6/E7 oncoprotein detection as a novel and promising triage strategy for managing HPV-positive women. Int J Cancer. 2019;144:34–42.
Schweizer J, Lu PS, Mahoney CW, Berard-Bergery M, Ho M, Ramasamy V, et al. Feasibility study of a human papillomavirus E6 oncoprotein test for diagnosis of cervical precancer and cancer. J Clin Microbiol. 2010;48:4646–8.
Sellors JW, Schweizer JG, Lu PS, Liu B, Weigl BH, Cui JF, et al. Association of elevated E6 oncoprotein with grade of cervical neoplasia using PDZ interaction-mediated precipitation of E6. J Low Genit Trac Dis. 2011;15:169–76.
Shi WJ, Liu H, Wu D, Tang ZH, Shen YC, Guo L. E6/E7 proteins are potential markers for the screening and diagnosis of cervical pre-cancerous lesions and cervical cancer in a Chinese population. Oncol Lett. 2017;14:6251–8.
Torres KL, Mariño JM, Pires Rocha DA, de Mello MB, de Melo Farah HH, Reis RDS, et al. Self-sampling coupled to the detection of HPV 16 and 18 E6 protein: A promising option for detection of cervical malignancies in remote areas. PLoS One. 2018;13:e0201262.
Torres-Ibarra L, Lorincz AT, Wheeler CM, Cuzick J, Hernández-López R, Spiegelman D, et al. Adjunctive testing by cytology, p16/Ki-67 dual-stained cytology or HPV16/18 E6 oncoprotein for the management of HPV16/18 screen-positive women. Int J Cancer. 2021;148:2264–73.
Valdez M, Jeronimo J, Bansil P, Qiao YL, Zhao FH, Chen W, et al. Effectiveness of novel, lower cost molecular human papillomavirus-based tests for cervical cancer screening in rural China. Int J Cancer. 2016;138:1453–61.
Wu ZN, Yu LL, Lei XQ, Qin Y, Zhang X, Chen W, et al. The association between human papillomavirus 16, 18 DNA load and E6 protein expression in cervical intraepithelial neoplasia and cancer. J Clin Virol. 2018;108:6–11.
Yang YS, Smith-McCune K, Darragh TM, Lai Y, Lin JH, Chang TC, et al. Direct human papillomavirus E6 whole-cell enzyme-linked immunosorbent assay for objective measurement of E6 oncoproteins in cytology samples. Clin Vaccin Immunol. 2012;19:1474–9.
Yu LL, Jiang MY, Qu PP, Wu ZN, Sun PS, Xi MR, et al. Clinical evaluation of human papillomavirus 16/18 oncoprotein test for cervical cancer screening and HPV positive women triage. Int J Cancer. 2018;143:813–22.
Zhang Q, Dong L, Hu S, Feng R, Zhang X, Pan Q, et al. Risk stratification and long-term risk prediction of E6 oncoprotein in a prospective screening cohort in China. Int J Cancer. 2017;141:1110–9.
Zhang Y, Sun L, Zhao H, Zhang Y, Wang L, Ma Z, et al. Application of human papillomavirus 16/18 E6 protein detection for cervical cancer screening. Cancer Res Clin. 2017;29:164–7.
Zhao FH, Jeronimo J, Qiao YL, Schweizer J, Chen W, Valdez M, et al. An evaluation of novel, lower-cost molecular screening tests for human papillomavirus in rural China. Cancer Prev Res. 2013;6:938–48.
Shukla S, Mahata S, Shishodia G, Pande S, Verma G, Hedau S, et al. Physical state & copy number of high risk human papillomavirus type 16 DNA in progression of cervical cancer. Indian J Med Res. 2014;139:531–43.
Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–84.
Acknowledgements
We thank Teresa Lee (knowledge manager, International Agency for Research on Cancer Lyon France) for her support regarding the research question building in all databases. We also thank Karima Bendeddouche for her support with formatting and submission process. Where authors are identified as personnel of the International Agency for Research on Cancer or World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer or World Health Organization.
Funding
This work was funded by the International Agency for Research on Cancer; The US National Cancer Centre for Global Health and the Horizon 2020 Framework Programme for Research and Innovation of the European Commission through the RISCC Network (Grant No. 847845). Sciensano, the employer of MA, received funding to conduct systematic reviews from the European Society of Gynaecologic Oncology.
Author information
Authors and Affiliations
Contributions
LD and M. Almonte conceived the study and protocol. LD developed the clinical question and defined the PICOS components with revision from M. Arbyn. LD developed the data extraction forms. LD and IJ extracted the data. MLR reviewed the data extraction. LZ extracted data from a Chinese paper with no English version. VNN trained LD on the metadta function and helped in designing the forest plots. LD conducted the statistical analyses with support from JV. LD wrote the report. A Baena provided guidance on statistical analyses. M. Gunter reviewed the manuscript. M. Almonte and M. Arbyn trained LD in meta-analytical methods and critically reviewed the report.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41416_2023_2490_MOESM1_ESM.docx
Supplementary materials to the manuscript entitled "Accuracy of HPV E6/E7 oncoprotein tests to detect high-grade cervical lesions: a systematic literature review and meta-analysis
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Downham, L., Jaafar, I., Rol, M.L. et al. Accuracy of HPV E6/E7 oncoprotein tests to detect high-grade cervical lesions: a systematic literature review and meta-analysis. Br J Cancer 130, 517–525 (2024). https://doi.org/10.1038/s41416-023-02490-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02490-w