Abstract
To investigate the chemical biodiversity of biologically active cyclic hexapeptides in the metabolites from microorganisms, the fermentation broth of Streptomyces alboflavus 313 was analyzed using HPLC, equipped with a diode array detector (DAD), coupled with ESI tandem MS (HPLC-DAD-ESI-MS/MS). In the mass spectra of cyclic hexapeptides, predominant ions [M+H]+, as well as [M-18+H]+, [M-28+H]+ and [M+Na]+, were observed and used to determine the molecular masses, while fragmentation reactions of [M+H]+ were recorded to provide information on the contents of amino acids and their linkage sequence. Based on the fragmentation patterns and comparison with standards, 15 pyrroloindoline-containing natural cyclic hexapeptides, cp01–15, were identified from the microorganism and six of these are reported for the first time.
Similar content being viewed by others
Introduction
Natural peptides are the largest chemical group of antibiotics produced by microorganisms, in which cyclopeptides have attracted more attention owing to their chemical diversity and impressive bioactivities.1 In our previous investigation of natural products from microorganisms for drug discovery, nine pyrroloindoline-containing cyclic hexapeptides, NW-G01, NW-G03 and NW-G05–NW-G11, were discovered in the fermentation broth of Streptomyces alboflavus 313.2, 3, 4, 5, 6, 7 They are structurally related to himastatin and chloptosin (Figure 1), two cyclic hexapeptides antibiotics produced by strains of Streptomyces. They all contain the common rare amino acids, piperazine-3-carboxylic acid and pyrroloindoline derivative, especially for chloptosin, it has the identical Trp-Val-PA3-PA2 moiety in its structure with that of NW-series cyclic hexapeptides. The largest difference between them is that the NW-series antibiotics are monomeric cyclohexapeptides, whereas himastatin and chloptosin are dimeric cyclohexapeptides. In terms of bioactivity, himastatin and chloptosin exhibit impressive antimicrobial activity against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus, whereas they were inactive against Gram-negative bacteria. In addition, they are also cytotoxic to mammalian tumor cell lines in vitro. The results of our investigation indicated that NW-series cyclohexapeptides bears similar antibacterial activity with those of himastatin and chloptosin. Benefiting from more analogs, a preliminary investigation on the relationship between structure and activity of these cyclopeptides has been carried out in our laboratory.
Attracted by their intriguing chemical structure and a medicinally relevant biological activity profile, himastatin, chloptosin and NW-G01 have been successfully synthesized in recent years.8, 9, 10, 11, 12 But these synthetic routes have only scientific value; large-scale fermentation is the only economically feasible way for mass production. To prompt development of new antibiotic and antitumor agents, scientists at Bristol Myers Squibb had achieved the optimization with ATCC 536653, the producer of himastatin, and the active compound could be obtained in scales adequate for sustaining chemical and biological investigation.8 Our previous investigation has indicated the chemical diversity of cyclopeptides in the metabolites of S. alboflavus 313, and the antibacterial activity of different analogs varied in a large extent. For drug discovery, screening for the best potential candidate from the microorganism is a valuable work.
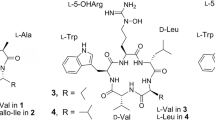
HPLC coupled with tandem MS (MS/MS) is a powerful technique that is amenable to analyze complex natural products. In recent years, this hybrid technique has also been used for the fast identification of natural peptides.13, 14, 15, 16, 17 To investigate the chemical biodiversity of biologically active cyclic hexapeptides in the metabolites from microorganism, we reported here the identification of pyrroloindoline-containing cyclic hexapeptides in the fermentation broth of S. alboflavus 313 by HPLC-diode array detector (DAD)-ESI-MS/MS. All NW-series cyclic hexapeptides consist of N-methylalanine (or alanine), valine (or leucine), chlorinated (or non-chlorinated) tryptophan derivative (Trp) and three piperazic acids (Pas; or its derivatives), their structures are shown in Figure 2. The target compounds were recognized first by the characteristic UV absorption of pyrroloindoline moieties and then were determined by the MS/MS experiments. As a result, 15 natural pyrroloindoline-containing cyclic hexapeptides were identified from the metabolites of this microorganism and six of them are reported for the first time.
Results and discussion
All NW-series cyclic hexapeptides bearing the same pyrroloindoline moiety showed the characteristic absorption at 230 and 300 nm. The UV absorption could be used to recognize the target compounds and the congeners. A characteristic UV spectrum obtained by DAD was shown in Figure 3. A representative UV (230 nm) chromatogram and total ions chromatogram of the cyclic hexapeptides in the extract of S. alboflavus 313 are shown in Figure 4. The target cyclic hexapeptides recorded at retention times from 8.69–27.17 min are designated as cp01 through cp15. Except for the dominant [M+H]+, [M+Na]+ ions also appeared in the mass spectra, thus the molecular mass of targets could be unambiguously determined. Structures for these cyclic hexapeptides were elucidated based on the fragmentation reactions of the [M+H]+ ions and are presented in the following sections.
Structural characterization of cp02, cp03, cp10, cp11 and cp13–15
Cp02, cp03, cp10, cp11, cp13–15 could unambiguously be identified as NW-G09, NW-G05, NW-G01, NW-G07, NW-G10, NW-G06 and NW-G08, respectively, based on comparison of the retention times with those of authentic reference substances and on the fragmentation pathways observed in the MS/MS experiments. For convenience of discussion, the amino acid residues constituting the peptide were labeled as the capital letters of their names; for example, valine (Val), N-methylalanine (Ala) and Trp derivatives (Trp) were labeled as V, A and T, respectively, and three molecules of PAs were coded as P1, P2 and P3, according to their positions in the structure. The fragmentations of these authenticated compounds are discussed in detail below (see for the case of cp10).
The full-scan mass spectrum of cp10 (NW-G01) contains [M+H]+ and [M+Na]+ ions at m/z 757 and 779, respectively, and the relative abundance (∼1/3) of the isotope peaks observed at m/z 759 and 781 indicated the presence of one chlorine atom (Figure 5). As shown in Figure 5, fragmentation of the [M+H]+ ion (m/z 757) yielded a series of product ions at m/z 739, 729, 672, 658, 533, 448, 409, 349 and 225, respectively. The predominant ion observed at m/z 739 was the product of water-loss from the moiety of chlorinated pyrroloindoline, for the only hydroxyl group was substituted at C3a-position of the Trp derivative. The ion at m/z 729 was 28 Da lower than the m/z value of [M+H]+, it might attribute to the loss of carbon monoxide (CO). Analyzing it’s product ions by using MS/MS/MS technique, the fragment of AP1TV was observed at m/z 533 in the spectrum, whereas the fragments containing P2P3, such as P1TVP3P2, P3P2AP1T, VP2P3A and P2P3 were not present; thus it could be determined that the CO was lost from the moiety of P2P3. Other ions observed in the collision induced dissociation (CID) spectrum were primarily produced from the cleavage of the peptide bonds. Careful comparison of the fragments with the structure of cp10, the ions at m/z 672, 658, 533, 448, 409, 349 and 225 were assigned to P1TVP3P2, P3P2AP1T, AP1TV, P1TV, VP2P3A, P1T and P2P3, respectively. The fragmentation pathway of cp10 was illustrated as Scheme 1, and the characteristic fragments were listed in Table 1.
Cp02, cp03, cp11, cp13–15 (NW-G09, NW-G05, NW-G07, NW-G10, NW-G06 and NW-G08) are structural similar to cp10. Compared with cp10, the only difference between cp02 was the proton at C5-position of Trp derivative in cp10, replaced by hydroxyl group; for cp03, N-methylalanine was replaced by alanine; for cp11 and cp14, PA2 were replaced by 5-methoxy-2,3-dihydropyridazine-3-carboxylic acid and 5-methoxy-2,3,4,5-tetra hydropyridazine-3-carboxylic acid; for cp13, N-methylalanine was replaced by alanine and PA2 was replaced by 5-methoxy-2,3-dihydropyridazine-3-carboxylic acid; for cp15, Val in cp10 was replaced by leucine, respectively. Except for the minor difference of individual amino acid residues, the sequence linkages of the amino acids in these cyclopeptides are identical with that of cp10. The characteristic fragments observed in their CID spectra could be elucidated based on their m/z values, and the amino acids contents of important fragments were listed in Table 1.
Except for the predominant [M+H]+ and [M+Na]+, the [M-28+H]+ and [M-18+H]+ ions were also observed in the full-scan mass spectra of these cyclopeptides, and they were identical to those ions produced from the neutral loss of water [M-18+H]+ or CO [M-28+H]+ in the CID spectra of all tested cyclic hexapeptides by comparing their product ions, so the characteristic ion peaks could be used to recognize this type of antibiotics.
Based on the above analysis for the authenticated compounds cp02, cp03, cp10, cp11, cp13–15, all NW-series cyclic hexapeptides showed very similar fragmentation patterns, it was consistent with their structural similarities. Using the proposed fragmentation pathway, the relative structure of the unknown cyclopeptide analogs in the metabolites of S. alboflavus 313 could be conveniently elucidated.
Structural characterization of cp01 and cp04
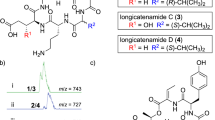
In the full-scan mass spectrum of cp04, the observed ions at m/z 695, 705, 723 and 745 could be readily deduced as [M-28+H]+, [M-18+H]+, [M+H]+ and [M+Na]+, the relative abundance of the isotope peaks revealed that there was no-chlorine atom in the structure. Because NW-G01 contains one chlorine atom, and cp04 is non-chlorinated, as well as the MW of NW-G01 (756) is 34 Da more than that of cp04 (722), so we hypothesized that these two compounds bear identical scaffold, except the proton at C6-position of the Trp was not replaced by chlorine atom in cp04. To test this hypothesis, their CID spectra were compared carefully. In the CID spectrum of NW-G01, the Trp-containing fragments P1TVP3P2, P3P2AP1T, AP1TV, P1TV and P1T were observed at m/z 672, 658, 533, 448 and 349, respectively, the corresponding ions (34 Da less) of cp04 appeared at m/z 638, 624, 499, 414 and 315 as expected. Meanwhile, the common fragments at m/z 409 and 225 were present in the CID spectra of NW-G01 and cp04, and they were deduced as VP3P2A and P2P3, respectively (see Supplementary Figure S1; Supplementary Scheme S1). Thus the structure of cp04 could be determined as a non-chlorinated derivative of NW-G01 (Figure 6). The characteristic product ions were tabulated in Table 1.
Subsequently, observed ions at m/z 693, 703, 721 and 743 could be readily deduced as [M-28+H]+, [M-18+H]+, [M+H]+ and [M+Na]+ in the full-scan mass spectrum of cp01. The relative abundance of the isotope peaks revealed that there was no-chlorine atom in the structure. The MW of cp01 (720) was 2 Da lower than that of cp04 (722), which suggested that one amino acid residue in cp01 was dehydrogenated. Compared its CID spectrum to that of cp04, the PA2-containing fragments P1TVP3P2, P3P2AP1T, VP3P2A and P1AP2 were observed at m/z 636, 622, 407 and 308, each were 2 Da lower than its corresponding ions of cp04, as well as the common ions AP1TV and P1T were observed at m/z 499 and 315 in their CID spectra, respectively. Thus, it could be concluded that the only difference between their structures was that one PA2 in cp01 was replaced by 2,3,4,5-tetrahydropyridazine-3-carboxylic acid, for this unsaturated heterocyclic carboxylic acid was widely distributed in the analogs reported previously (Figure 6).
Cp01 and cp04 are two novel natural peptides which have never been reported.
Structural characterization of cp05, cp06, cp08 and cp09
Cp05 and cp08 were detected in the selected ion chromatogram at 13.32 and 15.94 min, and their [M+H]+ were observed at m/z 755 (see Supplementary Figure S2). By comparison of the retention time with that of authentic reference substance and on the fragmentation pathways observed in the MS/MS experiments, cp05 could be unambiguously determined as NW-G03. The only difference between the structures of NW-G03 and NW-G01 was that PA3 in the former was replaced by 2,3,4,5-tetrahydropyridazine-3-carboxylic acid, and NW-G03 was the first discovered analog containing unsatruated PA moiety from S. alboflavus 313.
The structure of cp08 was speculated by comparing the characteristic fragments in its CID spectrum with those of cp05. The common characteristic neutral loss fragments, [M-18+H]+ (m/z 737) and [M-28+H]+ (m/z 727), as well as the similar UV spectra, indicated that cp08 had identical skeleton as cp05. The common ion m/z 656 that was assigned to P3P2AP1T in the fragmentation pathway of cp05 revealed that their amino acid contents were also similar, and the only difference might lie in the linkage sequence of PA3, PA2, Ala, PA1 and Trp. In the CID spectrum of cp05, m/z 349 and 533 were determined as the moieties of P1T and AP1TV, respectively, whereas the corresponding fragments were observed at m/z 347 and 531 in the CID spectrum of cp08, we hypothesized that PA1 was 2,3,4,5-tetrahydropyridazine-3-carboxylic acid in cp08 by analyzing structures of the analogs (Supplementary Figure S3; Supplementary Scheme S2). Thus, the possible structure of cp08 could be proposed, the only structure difference between this pair of isomers was the position of unsaturated PA (Figure 6). This hypothesis could also be well testified by the MS/MS experiment.
Likewise, cp06 and cp09 were another pair of isomers detected in the selected ion chromatogram, and their [M+H]+ were observed at m/z 753. It could be readily determined that there were two molecules of 2,3,4,5-tetrahydropyridazine-3-carboxylic acid in their structures by comparison of their CID spectra with those of other authenticated analogs. For cp06, the presence of m/z 533 indicated that it had the identical AP1TV moiety with NW-G01 and NW-G03, m/z 306, which was assigned to P3P2A, was 2 Da lower than the corresponding fragment (m/z 308) of NW-G03; PA2 could be determined as unsaturated PA (see Supplementary Figure S4; Supplementary Scheme S3). Thus, the structure of cp06 was proposed, in which PA2 and PA3 were 2,3,4,5-tetrahydropyridazine-3-carboxylic acids. For cp09, the fragments observed at m/z 531 and 223 were assigned to AP1TV and P2P3, respectively, the former was 2 Da less and the later was 2 Da more than the corresponding ions of cp06, it indicated that PA1 in cp09 was 2,3,4,5-tetrahydropyridazine-3-carboxylic acids (Figure 6).
Cp06, cp08 and cp09 are three novel natural peptides which have never been reported.
Structural characterization of cp07 and cp12
Cp07 and cp12 were a pair of isomers and their [M+H]+ were observed at m/z 781. Cp07 could be determined as NW-G11 by comparison of its retention time and the characteristic fragments of MS/MS experiment with those of authentic reference substance. For NW-G11, PA2 was 5-methoxy-2,3-dihydropyridazine-3-carboxylic acid, the conjugated heterocycle carboxylic acid is also present in the structures of NW-G06 and NW-G10. The structure of cp12 was speculated based on its characteristic fragments in CID spectrum and by comparing those with the corresponding ions of cp07. For cp07, AP1TV and P2P3 were observed at m/z 533 and 249, whereas the corresponding fragments of cp12 appeared at m/z 531 and 251, respectively (Supplementary Figure S5; Supplementary Scheme S4). As we know, PA3 in cp07 was 2,3,4,5-tetrahydropyridazine-3-carboxylic acid, the observed values indicated that PA1 in cp12 was the unsaturated PA, so the structure of cp12 was proposed, and the only difference between this isomers was the position of the unsaturated PA (Figure 6). The results of MS/MS experiments indicated that the proposed structure of cp12 was correct, and it was a novel natural peptide that has never been reported.
Experimental Procedure
Materials
Authenticated cyclic hexapeptides, NW-G01, NW-G03 and NW-G05–NW-G11, were obtained from the fermentation broth of S. alboflavus 313 by standard chromatographic procedures, and their structures were elucidated on the basis of extensive spectroscopic analysis. Acetonitrile (chromatographic grade) used for HPLC analysis was purchased from Tedia Chemical (Fairfield, OH, USA). Acetic acid, and the solvents (methanol and ethyl acetate) used in the extraction and column chromatography were analytical grade. The water used in HPLC analysis was purified on a Milli-Q system (Millipore, Bedford, MA, USA). Chromatography silica gel (200–300 mesh) was supplied by Qingdao Marine Chemical (Qingdao, China). The producing strain S. alboflavus 313 was isolated from a soil sample collected in Qinling Mountain, Shaanxi Province, China, its identification and fermentation have been described in our previous publications.2
Sample preparation
The fermentation broth (1 L) of S. alboflavus 313 was filtered with cheesecloth to separate the medium and culture liquid. The filtrate was extracted with ethyl acetate for three times (200 ml each time), and then the organic layers were combined and evaporated in vacuum. The concentrate was subjected to a silica gel column (10 g) and eluted with the mixture (50 ml) of EtOAc-MeOH at the ratio of 75:25 (v/v). The eluate was concentrated in vacuum and then dissolved in 10 ml acetonitrile for HPLC analysis.
Chromatography
A Thermo Surveyor HPLC system (Thermo, San Jose, CA, USA) with a Quat Pump, DAD, a Rheodyne 7725i injector with 10-μL sample loop, along with an Xcalibur software (Thermo, San Jose, CA, USA), was used. A reversed-phase C18 column (Hypersil Gold, 150 mm × 2.1 mm i.d., 5 μm) was used for separation; the column was kept at room temperature. The mobile phase consisted of acetonitrile (45%, v/v) and 0.1% aqueous acetic acid (55%, v/v) to start, followed by a linear gradient from 45–75% (v/v) acetonitrile over 40 min. During the next 10 min, the percentage of acetonitrile was maintained at 75%. The flow rate was 0.2 ml per min with UV detection at the range of 200–400 nm by photodiode array detection.
Mass spectrometry
HPLC/ESI-MS/MS analyses were performed using the Thermo Surveyor HPLC system described above combined with a Thermo Finnigan LCQ Advantage Max ion-trap mass spectrometer (Thermo Fisher Co., San Jose, CA, USA) equipped with an ESI interface. Instrument control and data acquisition were performed using Xcalibur 1.4 software. The capillary temperature was 300 °C, and the ESI needle voltage was always set at 4.5 kV. Nitrogen was used as sheath and auxiliary gas, the flow rate of them was set at 75 and 10 (arbitrary unit), respectively. Ultrahigh-purity helium was used as the damping and collision gas at a pressure of 0.133 Pa. A data-dependent acquisition was used in the LC-MS/MS experiments. The collision energy for MS/MS was adjusted to 35% and the isolation width of precursor ions was 2.5 mass units.
Conclusions
Although definitive structural proof for complex molecules can only be achieved through the use of many other sophisticated techniques, such as high resolution NMR and single crystal X-ray analysis, recently developed MS techniques have proven to be very useful in the detection and identification of a large variety of natural products.18, 19
In the current investigation, a reliable and simple analytical strategy based on a HPLC-DAD-ESI-MS/MS has been demonstrated to be efficient for the rapid detection of low-abundance cyclic hexapeptides in the fermentation broth of S. alboflavus 313. The molecular scaffolds were first recognized by the characteristic absorption of pyrroloindoline moieties at 230 and 300 nm obtained by DAD, and their molecular weights were determined from the [M+H]+, [M+Na]+, [M-18+H]+ and [M-28+H]+ ions observed in the full screen mass spectra, and their relative structures were established based on the proposed fragmentation pathway finally.

Fragmentation pathways proposed of cp10 (NW-G01). A full color version of this figure is available at The Journal of Antibiotics journal online.
References
Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 58, 1–26 (2005).
Guo, Z. Y., Shen, L., Ji, Z. Q., Huang, L. Z. & Wu, W. J. NW-G01, a novel cyclic hexadepsipeptide antibiotic, produced by Streptomyces alboflavus 313: I. Taxonomy, fermentation, isolation, physicochemical properties and antibacterial activities NW-G01 produced by Streptomyces alboflavus 313. J. Antibiot. 62, 201–205 (2009).
Guo, Z. Y. et al. NW-G01, a novel cyclic hexapeptide antibiotic, produced by Streptomyces alboflavus 313, II. Structural elucidation. J. Antibiot. 63, 231–235 (2010).
Guo, Z. Y. et al. NW-G03, a related cyclic hexapeptide compound of NW-G01, produced by Streptomyces alboflavus 313. J. Antibiot. 64, 789–794 (2011).
Ji, Z. Q., Qiao, G., Wei, S. P., Fan, L. X. & Wu, W. J. Isolation and characterization of two novel antibacterial cyclic hexapeptides from Streptomyces alboflavus 313. Chem. Biodivers. 9, 1567–1578 (2012).
Ji, Z. Q., Wei, S. P., Fan, L. X. & Wu, W. J. Three novel cyclic hexapeptides from Streptomyces alboflavus 313 and their antibacterial activity. Eur. J. Med. Chem. 50, 296–303 (2012).
Wei, S. P., Ji, Z. Q., Fan, L. X. & Wu, W. J. Two piperazic acid-containing cyclic hexapeptides from Streptomyces alboflavus 313. Amino Acids 43, 2191–2198 (2012).
Kamenecka, T. M. & Danishefsky, S. J. Studies in the total synthesis of himastatin: a revision of the stereochemical assignment. Angew. Chem. Int. Ed. 37, 2993–2995 (1998).
Kamenecka, T. M. & Danishefsky, S. J. Total synthesis of himastatin: confirmation of the revised stereostructure. Angew. Chem. Int. Ed. 37, 2995–2998 (1998).
Oelke, A. J. et al. Total synthesis of chloptosin: A dimeric cyclohexapeptide. Chem.-Eur. J. 17, 4183–4194 (2011).
Oelke, A. J., France, D. J., Hofmann, T., Wuitschik, G. & Ley, S. V. Total synthesis of chloptosin. Angew. Chem. Int. Ed. 49, 6139–6142 (2010).
Shibahara, S., Matsubara, T., Takahashi, K., Ishihara, J. & Hatakeyama, S. Total synthesis of NW-G01, a cyclic hexapeptide antibiotic, and 34-epi-NW-G01. Org. Lett. 13, 4700–4703 (2011).
Mohimani, H. et al. Multiplex de novo sequencing of peptide antibiotics. J. Comput. Biol. 18, 1371–1381 (2011).
Mohimani, H. et al. Sequencing cyclic peptides by multistage mass spectrometry. Proteomics 11, 3642–3650 (2011).
Malfanova, N., Franzil, L., Lugtenberg, B., Chebotar, V. & Ongena, M. Cyclic lipopeptide profile of the plant-beneficial endophytic bacterium Bacillus subtilis HC8. Arch. Microbiol. 194, 893–899 (2012).
Yu, Z. P. et al. Primary and secondary structure of novel ACE-inhibitory peptides from egg white protein. Food Chem. 133, 315–322 (2012).
Catiau, L., Delval-Dubois, V., Guillochon, D. & Nedjar-Arroume, N. Characterization and identification of a chymotryptic hydrolysate of alpha-lactalbumin stimulating cholecystokinin release in STC-1 Cells. Appl. Biochem. Biotechnol. 165, 1264–1273 (2011).
Niessen, W. M. A. Advances in instrumentation in liquid chromatography mass spectrometry and related liquid-introduction techniques. J. Chromatogr. A 794, 407–435 (1998).
Niessen, W. M. A. State-of-the-art in liquid chromatography-mass spectrometry. J. Chromatogr. A 856, 179–197 (1999).
Acknowledgements
This study was supported part by the grant of the National Natural Science Foundation of China (No.30971935), Program for New Century Excellent Talents in University from Education Ministry of China and Postdoctoral Science Foundation of China (20100471644 and 201104683).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Ji, Z., Xu, N., Gang, Q. et al. Identification of pyrroloindoline-containing cyclic hexapeptides in the metabolites of Streptomyces alboflavus 313 by HPLC-DAD-ESI-MS/MS. J Antibiot 66, 265–271 (2013). https://doi.org/10.1038/ja.2013.7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.7