Abstract
Oxidative stress contributes to dysfunction of glial cells in the optic nerve head (ONH). However, the biological basis of the precise functional role of mitochondria in this dysfunction is not fully understood. Coenzyme Q10 (CoQ10), an essential cofactor of the electron transport chain and a potent antioxidant, acts by scavenging reactive oxygen species (ROS) for protecting neuronal cells against oxidative stress in many neurodegenerative diseases. Here, we tested whether hydrogen peroxide (100 μM H2O2)-induced oxidative stress alters the mitochondrial network, oxidative phosphorylation (OXPHOS) complex (Cx) expression and bioenergetics, as well as whether CoQ10 can ameliorate oxidative stress-mediated alterations in mitochondria of the ONH astrocytes in vitro. Oxidative stress triggered the activation of ONH astrocytes and the upregulation of superoxide dismutase 2 (SOD2) and heme oxygenase-1 (HO-1) protein expression in the ONH astrocytes. In contrast, CoQ10 not only prevented activation of ONH astrocytes but also significantly decreased SOD2 and HO-1 protein expression in the ONH astrocytes against oxidative stress. Further, CoQ10 prevented a significant loss of mitochondrial mass by increasing mitochondrial number and volume density and by preserving mitochondrial cristae structure, as well as promoted mitofilin and peroxisome-proliferator-activated receptor-γ coactivator-1 protein expression in the ONH astrocyte, suggesting an induction of mitochondrial biogenesis. Finally, oxidative stress triggered the upregulation of OXPHOS Cx protein expression, as well as reduction of cellular adeonsine triphosphate (ATP) production and increase of ROS generation in the ONH astocytes. However, CoQ10 preserved OXPHOS protein expression and cellular ATP production, as well as decreased ROS generation in the ONH astrocytes. On the basis of these observations, we suggest that oxidative stress-mediated mitochondrial dysfunction or alteration may be an important pathophysiological mechanism in the dysfunction of ONH astrocytes. CoQ10 may provide new therapeutic potentials and strategies for protecting ONH astrocytes against oxidative stress-mediated mitochondrial dysfunction or alteration in glaucoma and other optic neuropathies.
Similar content being viewed by others
Main
Impaired mitochondrial oxidative phosphorylation (OXPHOS) generates excessive reactive oxygen species (ROS), leading to mitochondrial damage, oxidative stress and apoptotic cell death.1, 2 Increasing evidence demonstrates that oxidative stress links to mitochondrial dysfunction in glaucomatous neurodegeneration.2, 3, 4, 5 A recent study reported that lamina cribrosa cells in the optic nerve head (ONH) from glaucomatous human patients showed increased ROS production, impaired mitochondrial function and elevated cytosolic Ca2+,3 suggesting that a cycle of oxidative stress, mitochondrial dysfunction and dysregulation of calcium homeostasis may contribute to the pathogenesis of ONH degeneration in glaucoma.3 Although alteration of antioxidant enzymes has been implicated in glaucomatous ONH astrocytes,6 it remains unknown whether oxidative stress alters the mitochondrial network and triggers bioenergetic dysfunction in the ONH astrocytes.
Glaucoma is the leading cause of irreversible blindness and affects 70 million people worldwide.7 Although elevated intraocular pressure (IOP) is an important risk factor for ONH degeneration and retinal ganglion cell (RGC) death in glaucoma, lowering IOP is not always effective for preserving visual function in patients.7 Alterations in astrocytes have been implicated as important pathophysiological mechanisms in glaucomatous ONH degeneration.8, 9, 10 However, the biological basis of the precise functional role of ONH astrocytes in glaucomatous neurodegeneration is not fully understood.
Since the first evidence of impaired mitochondrial respiration-mediated mitochondrial dysfunction in patients with primary open-angle glaucoma,4 we have found that mitochondrial dysfunction is associated with ONH degeneration and RGC death in a mouse model of glaucoma,11, 12, 13 suggesting a distinct mitochondrial dysfunction-mediated degenerative pathway in glaucomatous neurodegeneration. Growing evidence indicates that glaucomatous damage, including elevated IOP, contributes to dysfunction of glial cells or loss of astrocytic processes in the ONH of rodent models of glaucoma.10, 14 Regardless, the relationship between mitochondrial dysfunction and ONH astrocytes in glaucomatous neurodegeneration remain unknown.
Coenzyme Q10 (CoQ10), an essential cofactor of the electron transport chain, acts by maintaining the mitochondrial membrane potential, supporting adenosine triphosphate (ATP) synthesis and inhibiting ROS generation, thus protecting neuronal cells against oxidative stress in neurodegenerative diseases.15, 16, 17 Of note, the levels of CoQ10 in the human retina can decline by ∼40% with age,18 raising the possibility that this decline of CoQ10 may contribute to age-related ROS generation in glaucomatous ONH neurodegeneration. Previous studies demonstrated that CoQ10 protects retinal cells against oxidative stress in vitro and in vivo, as well as prevents retinal damage induced by acute IOP elevation or excitotoxicity in vivo.19, 20, 21, 22
Here, we tested whether hydrogen peroxide (H2O2)-induced oxidative stress alters the mitochondrial network, bioenergetics and OXPHOS complex (Cx) expression, and whether CoQ10 can ameliorate oxidative stress-mediated alterations in mitochondria of ONH astrocytes in vitro.
Results
CoQ10 ameliorates oxidative stress in the ONH astrocytes
Pure ONH astrocytes were generated from postnatal day 5 Sprague–Dawley rats using a modification of a previous protocol by Hernandez et al.23 As shown in Figure 1, the cells migrated from cultured ONH tissues were purified by vigorous shaking and serum deprivation (Figure 1a). To confirm whether purified cells were astrocytes, we performed Western blot and immunocytochemical analyses using markers for astrocytes, microglial cells and oligodendrocytes. We found that the purified cells were positive for glial fibrillary acidic protein (GFAP), a marker for astrocytes, but were negative for Iba1, a marker for microglial cells, and myelin basic protein (MBP), a marker for oligodendrocytes (Figure 1b). In contrast, the protein extract from rat ON tissues contained all four of markers. Immunocytochemistry showed a strong immunoreacitivity for GFAP in cultured ONH astrocytes (Figure 1b), suggesting that the purified cells from ONH tissues were astrocytes.
Characterization of cultured rat optic nerve head (ONH) astrocytes. (a) The diagram represents purification of ONH astrocytes from rats. (b) The protein extracts from ONH astrocytes were positive for GFAP, a marker for astrocytes but were negative for Iba1, a marker for microglial cells, and MBP, a maker for oligodendrocytes. The protein extract from rat optic nerve tissue was used for positive control. A representative image shows that GFAP immunoreactivity is present in the ONH astrocytes. GFAP, glial fibrillary acidic protein; MBP, myelin basic protein. Scale bar, 20 μm
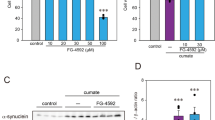
As reduction of oxidative stress has been proposed to be protective against glaucomatous damage,24, 25 we determined whether CoQ10 treatment prevents oxidative stress-mediated activation of ONH astrocytes in vitro using antibodies for GFAP, superoxide dismutase 2 (SOD2) and heme oxygenase-1 (HO-1). We observed that GFAP protein expression was significantly increased by 1.23±0.10-fold in the ONH astrocytes exposed to H2O2 (100 μM) compared with control cells exposed to vehicle. In contrast, CoQ10 significantly reduced GFAP protein expression by 0.97±0.08-fold in the ONH astrocytes exposed to H2O2 (Figure 2a). Intriguingly, we also found that SOD2 and HO-1 protein expression was significantly increased by 1.40±0.13- and 1.2±0.14-fold in the ONH astrocytes exposed to H2O2 compared with control cells, respectively. In contrast, CoQ10 significantly reduced SOD2 and HO-1 protein expression by 0.63±0.11- and 0.63±0.07-fold in the ONH astrocytes exposed to H2O2, respectively (Figure 2a). To determine whether oxidative stress alters SOD2 distribution and expression in the ONH astocytes, we performed immunocytochemistry in cultured ONH astrocytes using the antibody for SOD2. Consistently, we found that SOD2 immunoreactivity was present in the mitochondria of the ONH astrocytes. Moreover, we observed that ONH astrocytes exposed to H2O2 showed increase of SOD2 immunoreactivity in the mitochondria and cytoplasm compared with control cells. In contrast, CoQ10 decreased SOD2 immunoreactivity in the ONH astrocytes exposed to H2O2 (Figure 2b). These results indicate that CoQ10 prevents activation of ONH astrocytes by blocking oxidative stress.
CoQ10 ameliorates oxidative stress in ONH astrocytes. (a) GFAP, SOD2 and HO-1 protein expression were significantly increased in the ONH astrocytes exposed to H2O2 (100 μM) compared with vehicle-treated control ONH astrocytes. In contrast, CoQ10 significantly reduced GFAP, SOD2 and HO-1 protein expression compared with ONH astrocytes exposed to H2O2. Relative intensity of chemiluminescence for each protein band was normalized using actin. Values are mean±S.D. *P<0.05 compared with vehicle-treated control ONH astrocytes or H2O2-treated ONH astrocytes. (b) Representative images show that SOD2 immunoreactivity was present in mitochondria of the ONH astrocytes. Of note, SOD2 immunoreactivity was increased in the ONH astrocytes exposed to H2O2. However, CoQ10 decreased SOD2 immunoreactivity in the ONH astrocytes exposed to H2O2. GFAP, glial fibrillary acidic protein; SOD2, superoxide dismutase 2; HO-1, heme oxygenase-1; CoQ10, coenzyme Q10; H2O2, hydrogen peroxide. Scale bars, 10 μm
CoQ10 triggers mitochondrial biogenesis against oxidative stress
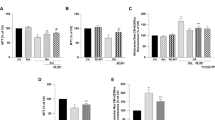
To determine whether oxidative stress triggers alteration of the intracellular mitochondrial network in ONH astrocytes and whether CoQ10 treatment inhibits this alteration in mitochondria against oxidative stress, the mitochondrial morphology of ONH astrocytes were assessed by MitoTracker Red (Invitrogen-Molecular Probes, Eugene, OR, USA) staining, a marker for mitochondria. Further, we quantified the aterations of mitochondrial number, length and volume density following exposure of H2O2 using transmission electron microscopy (TEM) analysis. Our results showed that control ONH astrocytes contained classic elongated tubular mitochondria. However, ONH astrocytes exposed to H2O2 contained small rounded mitochondria. Interestingly, ONH astrocytes pretreated with CoQ10 and exposed to H2O2 showed a partial preservation of mitochondrial morphology compared with the ONH astrocytes exposed to H2O2 (Figure 3). Of note, representative 2D images from TEM showed that ONH astrocytes exposed to oxidative stress produced fewer mitochondria. In good agreement with this result, quantitative analyses importantly showed that the number of mitochondria, normalized to the total area occupied by somas in each image, was significantly decreased in the ONH astrocytes exposed to H2O2 (0.15±0.01 μm2) compared with control ONH astrocytes (0.52±0.07 μm2; P<0.001; Figures 4a and b). In contrast, pretreatment of CoQ10 increased mitochondrial numbers to a lesser extent (0.26±0.03) compared with ONH astrocytes exposed to H2O2 (P<0.05; Figure 4b). In addition, mitochondrial volume density, defined as the volume occupied by mitochondria divided by the volume occupied by the cytoplasm in terms of a percentage, was decreased in the ONH astrocytes exposed to H2O2 (3.58±0.38%) compared with control ONH astrocytes (6.36±0.87%; P<0.01; Figures 4a and b). In contrast, pretreatment of CoQ10 significantly increased the mitochondrial volume density (5.84±0.75%) compared with ONH astrocytes exposed to H2O2 (P<0.05; Figure 4b). Interestingly, however, there was no difference in mitochondrial length among control, H2O2- and CoQ10/H2O2-treated ONH astrocytes (Figures 4a and b), indicating that mitochondrial biogenesis by CoQ10 did not come from increased mitochondrial fusion but rather by nascent mitochondria.
CoQ10 partially preserves mitochondrial structure of ONH astrocytes against oxidative stress-induced mitochondrial fission. The mitochondria of ONH astrocytes were stained with MitoTracker Red. Control ONH astrocytes exposed to vehicle contained classic elongated tubular mitochondria. In contrast, ONH astrocytes exposed to H2O2 contained small rounded mitochondria. However, ONH astrocytes pretreated with CoQ10 showed a partial preservation of mitochondrial morphology compared with the ONH astrocytes exposed to H2O2. CoQ10, coenzyme Q10; H2O2, hydrogen peroxide. Scale bars, 10 μm
CoQ10 triggers mitochondrial biogenesis in ONH astrocytes that counters oxidative stress. (a) The representative 2D images from TEM analysis showed that control ONH astrocytes exposed to vehicle contained classic elongated tubular mitochondria. However, ONH astrocytes exposed to H2O2 contained fewer mitochondria in the ONH astrocytes. Interestingly, ONH astrocytes pretreated with CoQ10 showed a greater number compared with the ONH astrocytes exposed to H2O2. (b) Quantitative analysis showed that mitochondrial number per area and volume density were significantly decreased in the ONH astrocytes exposed to H2O2. However, ONH astrocytes pretreated with CoQ10 had a significant increase in mitochondrial number and volume density compared with the ONH astrocytes exposed to H2O2. There was no difference in mitochondrial length among control, H2O2 and CoQ10/H2O2-treated ONH astrocytes. Values are mean±S.E.M. *P<0.05, **P<0.01 and ***P<0.001 compared with vehicle-treated control ONH astrocytes or H2O2-treated ONH astrocytes. Scale bar, 500 nm. (c) Mitofilin protein expression was significantly increased in the ONH astrocytes exposed to H2O2 compared with vehicle-treated control ONH astrocytes. In contrast, CoQ10 significantly increased mitofilin protein expression compared with ONH astrocytes exposed to H2O2. PGC-1α protein expression was significantly increased in the ONH astrocytes exposed to H2O2 compared with vehicle-treated control ONH astrocytes. Of interest, pretreatment of CoQ10 showed greater increase of PGC-1α protein expression in the ONH astrocytes exposed to H2O2. Relative intensity of chemiluminescence for each protein band was normalized using actin. Values are mean±S.D. (n=3). CoQ10, coenzyme Q10; H2O2, hydrogen peroxide
CoQ10 promotes mitofilin and PGC-1α protein expression against oxidative stress
The mitochondrial inner membrane protein mitofilin is critical for maintenance of mitochondrial cristae morphology and is reduced following oxidative stress.26, 27 To determine whether H2O2-induced oxidative stress triggers loss of mitofilin protein expression in the ONH astrocytes and whether CoQ10 treatment prevents this loss, we examined the protein expression level of mitofilin using western blot analysis. We found that oxidative stress triggered a significant loss of mitofilin protein expression by 0.36±0.04-fold in the ONH astrocytes (P<0.01; Figure 4c). In contrast, CoQ10 significantly promoted mitofilin protein expression by 0.59±0.02-fold in the ONH astrocytes exposed to H2O2 compared with H2O2-treated ONH astrocytes (P<0.05; Figure 4c). However, there was no difference of porin protein expression between H2O2 and CoQ10/H2O2-treated ONH astrocytes (Figure 4c), suggesting that CoQ10 could be protective in the mitochondria of ONH astrocytes by increasing the expression of mitofilin that in turn may protect mitochondrial cristae structure and ultimately OXPHOS capacity against oxidative stress. On the basis of increased mitochondrial mass by increasing mitofilin protein expression, we also determined whether CoQ10 triggers mitochondrial biogenesis using the antibody for peroxisome-proliferator-activated receptor-γ coactivator-1 (PGC-1α), a transcriptional coactivator and a mediator of mitochondrial biogenesis, in the ONH astrocytes exposed to H2O2. We found that oxidative stress significantly increased PGC-1α protein expression by 1.34±0.22-fold in the ONH astrocytes (P<0.05; Figure 4c). Of interest, CoQ10 showed greater increase of PGC-1α protein expression by 1.74±0.40-fold in the ONH astrocytes exposed to H2O2 (P<0.05; Figure 4c), suggesting that CoQ10 could also be protective in the ONH astrocytes by triggering mitochondrial biogenesis against oxidative stress.
To better visualize the cristae, we performed electron tomography, a technique that provides the highest resolution three-dimensional (3D) structure determination of mitochondria. We generated tomographic reconstructions of 15 control, 60 H2O2-exposed and 33 CoQ10/H2O2-treated mitochondria and noticed that the cristae appeared dilated in the H2O2-exposed mitochondria (Figure 5). Measurements of cristae widths in the mitochondrial volumes showed that control cristae were 32±5 nm (mean±S.D.) across (membranes included), CoQ10/H2O2-treated cristae were 31±6 nm across, yet H2O2-exposed cristae were 48±17 nm across, a value statistically different from control and CoQ10/H2O2-treatment values (P<0.001, Figure 5). The variation in cristae width was much greater in the H2O2-exposed mitochondria reflecting that not all the cristae were dilated. In comparison, the cristae widths in control and CoQ10/H2O2-treated mitochondria were more uniform (Figure 5). The abnormal cristae associated with oxidative stress are consistent with our finding of reduced ATP production. To further determine cristae abundance, we also performed measurment of mitochondrial cristae abundance. The parameter is cristae membrane surface area (SA) normalized to the outer membrane area, that is, cristae membrane SA divided by outer membrane SA that is dimensionless. Measurement of cristae abundance in the mitochondria showed that H2O2-exposed cristae significantly increased cristae abundance by 1.75±0.09 (n=5, mean±S.E.M.) compared with control cristae abundance by 0.95±0.26 (n=5; P<0.05, Figure 5). Pretreatment of CoQ10 decreased cristae abundance by 1.43±0.33 (n=5) in the ONH astrocytes exposed to H2O2; however, there was no statistical difference between H2O2 and CoQ10/H2O2-treated ONH astrocytes (Figure 5).
3D reconstruction of mitochondrial cristae in the ONH astrocytes. Oxidative stress dilated cristae that were prevented by CoQ10/H2O2 treatment. Electron tomography generated high-resolution, 3D reconstructions of control, H2O2-exposed and CoQ10/H2O2-treated mitochondria. Slices (1.4-nm thick) through the middle of electron microscopy tomographic volumes of mitochondria are shown on the left. Surface-rendered volumes of the segmented mitochondria provide information concerning shape and cristae architecture. The outer mitochondrial membrane is shown in blue (made translucent to better visualize the cristae) and cristae are in various colors. The long control mitochondrion has 46 cristae, the H2O2-exposed has 47 cristae distributed in the four mitochondria that are lined up and the CoQ10/H2O2-treated mitochondrion has 27 cristae. The mean of cristae widths is 50% greater in the H2O2-exposed mitochondria compared with the control and CoQ10 pretreatment samples. Scale bar, 250 nm (all panels). Values are mean ±S.E.M. *Significant at P<0.05 and ***Significant at P<0.001 compared with vehicle-treated control ONH astrocytes or H2O2-treated ONH astrocytes. Representative graphs show the measurement of cristae widths and abundance in the mitochondria. CoQ10, coenzyme Q10; H2O2, hydrogen peroxide
CoQ10 preserves OXPHOS Cx protein expression against oxidative stress
Pretreatment of CoQ10 has been proposed to have a beneficial effect in the treatment of OXPHOS disorders.28, 29, 30 To determine whether H2O2-induced oxidative stress alters OXPHOS Cx protein expression and whether CoQ10 preserves OXPHOS Cx protein expression in ONH astrocytes, we examined the protein expression level of OXPHOS Cxs using western blot analysis. As shown in Figure 6, H2O2-induced oxidative stress significantly increased OXPHOS Cx (Cx I and II) protein expression by 2.56±0.08-(P<0.001) and 1.93±0.09-fold (P<0.01) compared with control ONH astrocytes, respectively. However, there were no significant difference in OXPHOS Cx (Cx III–V) protein expression between control and H2O2-treated ONH astrocytes. We also found that pretreatment of CoQ10 significantly decreased OXPHOS Cx (Cx I, II and V) protein expression by 0.33±0.06- (P<0.001), 1.36±0.16- (P<0.01), and 0.62±0.07-fold (P<0.05), respectively, compared with ONH astrocytes exposed to H2O2, However, there was no significant difference in OXPHOS Cx (Cx III and IV) protein expression between H2O2- and CoQ10/H2O2-treated ONH astrocytes, indicating that CoQ10 may preserve OXPHOS Cx protein expression in the ONH astrocytes against oxidative stress.
CoQ10 preserves OXPHOS Cx protein expression against oxidative stress. In comparison with control ONH astrocytes, H2O2-induced oxidative stress significantly increased OXPHOS Cx (Cx I and II) protein expression in the ONH astrocytes. However, there was no significant difference in OXPHOS Cx (Cx III–V) protein expression. Pretreatment of CoQ10 significantly decreased OXPHOS Cx (Cx I, II and V) protein expression compared with ONH astrocytes exposed to H2O2. However, there was no significant difference in OXPHOS Cx (Cx III and IV) protein expression between H2O2- and CoQ10/H2O2-treated ONH astrocytes. Values are mean±S.D. *P<0.05, **P<0.01 and ***P<0.001 compared with vehicle-treated control ONH astrocytes or H2O2-treated ONH astrocytes. CoQ10, coenzyme Q10; H2O2, hydrogen peroxide; OXPHOS, oxidative phosphorylation; Cx, complex
CoQ10 prevents alteration of mitochondrial bioenergetics due to oxidative stress
To determine whether H2O2-induced oxidative stress alters mitochondrial bioenergetics in ONH astrocytes and whether CoQ10 treatment inhibits the alteration of mitochondrial bioenergetics, we measured cell viability, ROS generation and cellular ATP level. As shown in Figure 7, our findings show that H2O2-induced oxidative stress significantly decreased cell viability to 80.8±5.4% of the control value in the ONH astrocytes (P<0.05). However, pretreatment of CoQ10 significantly boosted cell viability to 92.6±4.6% in the ONH astrocytes exposed to H2O2 (P=0.001). In comparison with control ONH astrocytes, H2O2-induced oxidative stress significantly increased ROS generation by 7.82±0.81-fold over the control in the ONH astrocytes (P=0.01). In contrast, pretreatment of CoQ10 significantly decreased ROS generation to 4.67±0.83-fold of the control in the ONH astrocytes exposed to H2O2 (P<0.05). In addition, we found that H2O2-induced oxidative stress significantly decreased the cellular ATP level to 83.8±3.7% of the control value in the ONH astrocytes (P<0.01). However, pretreatment of CoQ10 nearly completely restored the cellular ATP level (96.9±4.3% of the control) in the ONH astrocytes exposed to H2O2 (P<0.05), suggesting that CoQ10 restores bioenergetic function in the ONH astrocytes to counter oxidative stress.
CoQ10 prevents the alteration of mitochondrial bioenergetics caused by oxidative stress. Pretreatment of CoQ10 significantly restored cell viability to 92.6±4.6% of the control level in the ONH astrocytes exposed to H2O2. In comparison with control ONH astrocytes, H2O2-induced oxidative stress significantly increased ROS generation in the ONH astrocytes. In contrast, pretreatment of CoQ10 significantly decreased ROS generation in the ONH astrocytes exposed to H2O2. In addition, H2O2-induced oxidative stress significantly decreased the cellular ATP level in the ONH astrocytes. However, pretreatment of CoQ10 significantly restored the cellular ATP level in the ONH astrocytes exposed to H2O2. Values are mean±S.D. *P<0.05 and **P<0.01 compared with vehicle-treated control ONH astrocytes or H2O2-treated ONH astrocytes. CoQ10, coenzyme Q10; H2O2, hydrogen peroxide
Discussion
We addressed the question of whether oxidative stress triggers mitochondria-mediated dysfunction in the ONH astrocytes because oxidative stress has been implicated as an important pathophysiological mechanisms in the pathogenesis of glaucoma.4, 9, 31, 32 It has been proposed that reduction of oxidative stress could be protective in glaucomatous ONH treatment24, 33, 34, 35 and that mitochondrial dysfunction may have a critical role in oxidative stress-mediated glaucomatous neurodegeneration.2, 4, 5, 36, 37 Interestingly, oxidative stress leads to antioxidant or immune responses in human ONH astrocytes.24, 32, 38 Support for our investigations came from a recent study demonstrating oxidative stress, mitochondrial dysfunction and Ca2+ overload in GFAP-negative human lamina cribrosa cells in the ONH from glaucoma donor eyes.3
CoQ10 is an attractive antioxidant and neurotherapeutic agent for us to test efficacy in glaucoma because published evidence supports its effectiveness against oxidative stress in many neurodegenerative diseases including Parkinson’ and Huntington’ diseases.15, 39, 40 Recent studies increasingly demonstrated that CoQ10 is neuroprotective in retinal cells in vivo and in vitro against pressure, oxidative stress, excitotoxicity, or apoptotic radiation.19, 21, 22, 41, 42 In the current study, we found that oxidative stress triggers the activation of ONH astrocytes as well as the upregulation of SOD2 and HO-1 protein expression in the ONH astrocytes. In contrast, CoQ10 not only prevents activation of ONH astrocytes but also significantly decreases SOD2 and HO-1 protein expression in the ONH astrocytes against oxidative stress. The upregulation of antioxidative enzymes such as SOD2 in astrocytes is accompanied by a higher resistance to oxidative stress induced by H2O2.43 In addition, SOD2 activity directly stabilizes mitochondrial transmembrane potential and calcium-buffering ability.44 Together with these findings, our results, therefore, raise the possibility that increasing SOD2 or HO-1 expression in the ONH astrocytes may contribute to compensatory endogenous antioxidant mechanisms that increase resistance or stabilization of mitochondria against oxidative stress. In addition, these results reflect that CoQ10 could be an important antioxidant for ameliorating oxidative stress-mediated dysfunction in the ONH astrocyte. We believe that studying the benefits of CoQ10-mediated blockade of oxidative stress or enhancement of antioxidant enzymes in the ONH astrocytes may be rewarding in many optic neuropathy including glaucoma.
We further addressed the relationship between oxidative stress and the mitochondrial network in the ONH astrocytes. We demonstrated for the first time that oxidative stress induced a significant loss of mitochondrial mass by decreasing mitochondrial number and volume density in the ONH astrocytes. However, CoQ10 treatment showed a partial inhibition of mitochondrial loss by increasing mitochondrial number and volume density in the ONH astrocytes, suggesting that CoQ10 could protect ONH astrocytes by triggering mitochondrial biogenesis. In agreement with these findings, we also observed that oxidative stress significantly reduced the expression level of mitofilin protein in the ONH astrocytes. However, there was no significant difference in porin expression among groups, suggesting that the observed increase in number of mitochondria did not produce significantly more outer membrane where porin resides or alternatively that porin was diluted upon mitochondrial biogenesis. Intriguingly, CoQ10 treatment partially increased mitofilin protein expression in the ONH astrocytes against oxidative stress. More importantly, we also found that CoQ10 treatment showed greater increase of PGC-1α protein expression in the ONH astrocytes to protect against oxidative stress.
Mitofilin is anchored to the mitochondrial inner membrane and controls mitochondrial cristae morphology.26 As a previous study suggests that mitofilin depletion decreases cellular proliferation and increases apoptosis, as well as triggers mitochondrial structural and functional abnormalities,26 it is possible that a significant loss of mitofilin protein expression by oxidative stress may result in structural and functional dysfunction of mitochondria in the ONH astrocytes. PGC-1α activates mitochondrial biogenesis and oxidative metabolism45, 46 as well as regulates the transcription target proteins such as nuclear-encoded respiratory Cx proteins and mitochondrial transcription factor A that has an important role in mitochondrial gene expression and mitochondrial DNA maintenance, and therefore is essential for OXPHOS-mediated ATP synthesis.45, 47, 48 Collectively, therefore, increasing mitofilin or PGC-1α protein expression by CoQ10 treatment also raises the intriguing possibility that mitofilin or PGC-1α can be a therapeutic target for protecting mitochondria or promoting mitochondrial biogenesis, especially cristae biogenesis or stablization of the ONH astrocytes to counter oxidative stress-mediated mitochondrial dysfunction. In addition, we found that oxidative stress significantly increased cristae widths and abundance in the mitochondria of the ONH astocytes. However, CoQ10 treatment significantly preserved cristae widths, but partially preserved cristae abundance in the mitochondria of the ONH astrocytes against oxidative stress, suggesting that CoQ10 can modulate mitochondrial cristae structure in the ONH astrocytes to protect against oxidative damage. Altogether, our findings suggest that CoQ10 may have a therapeutic potential for ameliorating oxidative stress-induced alteration of the mitochondrial network in the ONH astrocytes. Future studies are needed to address how CoQ10 directly promotes mitochondrial biogenesis and structural preservation in the ONH astrocytes against oxidative stress and whether oxidative stress-mediated mitofilin loss directly alters mitochondrial cristae morphology.
In the current study, we found that oxidative stress triggered the upregulation of OXPHOS Cxs (Cx I–IV) protein expression in the ONH astocytes. However, CoQ10 treatment preserved OXPHOS Cx protein expression including the ATP synthase against oxidative stress. CoQ10 regulates the mitochondrial OXPHOS system and prevents lipid peroxidation.49, 50 It has been demonstrated that a defective OXPHOS system may be induced by an insufficient fuel supply, defective electron transport chain enzymes (Cx I–IV) or lack of the electron carrier CoQ10.49 In addition, CoQ10 treatment significantly increases ATP synthetic capacity in lymphocytes from patients as well as in cultured control lymphocytes in vitro,28 suggesting that CoQ10 may have a beneficial effect in the treatment of OXPHOS disorders.28, 29 Although further investigations are needed, it is possible that oxidative stress may induce CoQ10 deficiency in ONH astrocytes, triggering alteration of the OXPHOS system. Further, our findings showed that CoQ10-mediated partial preservation of mitochondrial cristae abundance supports CoQ10-mediated increase of certain OXPHOS Cxs against oxidative stress. Thus, our results reflect that the upregulation of OXPHOS Cx protein expression may contribute to a critical endogenous defense mechanism for protecting mitochondria of the ONH astrocytes against oxidative stress.
Oxidative stress-mediated ROS alters OXPHOS function.51, 52 This alteration of mitochondrial OXPHOS Cxs by oxidative stress in turn leads to reduction of ATP production and increase of ROS generation, consistent with our results.2 Further, increased ROS generation results in mitochondrial bioenergetic dysfunction in many neurodegenerative diseases,53, 54 suggesting an oxidative stress-mediated mitochondrial vicious cycle involved in neurodegeneration that includes oxidative stress, ROS production, OXPHOS alteration and mitochondrial bioenergetic dysfunction. In the current study, we found that oxidative stress triggers an excessive increase of ROS generation, reduction of cellular ATP level and subsequent decrease of cell viability in the ONH astrocytes. Of note, we found that CoQ10 treatment decreases ROS generation, increases cellular ATP production, and promotes cell viability in ONH astrocytes. It was shown that CoQ10 improves mitochondrial bioenergetic parameters such as oxygen consumption rate, mitochondrial transmembrane potential and ATP synthesis in cultured T62 and H9c2 cell lines consistent with its role in the mitochondrial respiratory chain.55 Moreover, a recent study demonstrated that CoQ10 deficiency is associated with oxidative stress, increased ROS production, defective ATP synthesis and cell death.56 Our findings also showed that oxidative stress triggered abnormal cristae morphology such as cristae dilation in the mitochondria of the ONH astroctyes. Moreover, because activation of PGC-1α correlates with increased cellular energy demand,57 a significant increase of PGC-1α may reflect an increased energy demand from the mitochondria of ONH astrocytes against oxidative stress. This correlates with our findings of reduced ATP production. Altogether, these findings strongly suggest that CoQ10 supplementation may have therapeutic potential for ameliorating a mitochondrial vicious cycle involved in ONH degeneration that includes oxidative stress, OXPHOS dysfunction, ROS generation and bioenergetics impairment in the ONH astrocytes during glaucomatous neurodegeneration. Finally, we believe that it would be useful to measure the origin of the energy defect by pharmacologically inhibiting glycolytic and mitochondrial pathways in the future studies.
On the basis of these observations, we suggest that oxidative stress-mediated mitochondrial dysfunction or alteration may be important pathophysiological mechanisms in the dysfunction of ONH astrocytes. Therefore, treatment with CoQ10 may be neuroprotective and protect ONH astrocytes against oxidative stress-mediated mitochondrial dysfunction or alteration in glaucoma and other optic neuropathies.
Materials and Methods
Animals
Pregnant Sprague–Dawley rats (250–300 g in weight; Harlan Laboratories, Indianapolis, IN, USA) were housed in covered cages, fed with a standard rodent diet ad libitum, and kept on a 12-h light/12-h dark cycle. All procedures concerning animals were in accordance with the ARVO Statement for the Use of Animals in Ophthalmic Vision Research and under protocols approved by institutional IACUC committees at the University of California, San Diego.
Isolation and culture of rat ONH astrocytes
Primary rat ONH astrocyte cultures were modified and derived as previously described.23 After euthanizing with CO2 gas, 10–20 pieces of ONH tissue were dissected from postnatal day 5 Sprague–Dawley rats and transferred to a 35-mm petri dish with 2 ml 0.2% bovine serum albumin (BSA)/Dulbecco’s phosphate-buffered saline (DPBS). Under a dissecting microscope, pigmented and remnant non-neuronal tissues such as sclera and other neighboring tissues were removed, and the ONH tissue was identified and dissected using a microscissor and sharp blade. The ONH tissue was minced and transferred to a 60-mm petri dish and conditioned with growth medium: GlutaMAX/Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, La Jolla, CA, USA), and prewarmed in a humidified atmosphere 5% CO2 incubator at 37 °C. After incubation for 10 days, the ONH explants were removed by 70-μm cell strainers (BD, Sparks, MD, USA). The cells that were grown from the ONH explants were plated in a 100-mm petri dish and incubated for 3 days at 37 °C. For further purification of the ONH astrocytes, the growth medium was changed to serum-deprived medium and the culture dish was shaken for 24 h at room temperature. After removing nonadherent cells, the adherent ONH astrocytes were collected, centrifuged and replated on poly-L-lysine-coated culture dishes. The homogeneity of ONH astrocyte cultures was evaluated by western blot analysis or immunohistochemistry as below.
Pharmacological treatment
Three groups of rat ONH astrocytes were studied following treatment of H2O2 or CoQ10: a group treated with vehicle (chloroform; Sigma, St. Louis, MO, USA), a group treated with 100 μM H2O2 (Sigma), and a group treated with CoQ10 (50 μg/ml in chloroform, Sigma) and 100 μM H2O2 (Sigma). H2O2 was treated in 1% FBS/DMEM/Ham’s F12 50/50 (DMEM/F12) for 1 h in a 5% CO2 incubator at 37 °C. CoQ10 was pre-incubated with 1% FBS/DMEM/F12 for 24 h and treated with H2O2 with DMEM/F12 for 1 h in a 5% CO2 incubator at 37 °C.
Western blot analysis
Rat ONH astrocytes were lysed with lysis buffer (20 mM HEPES pH 7.0, 1% TritonX-100, 10% glycerol, 2 mM EGTA, 1 mM EDTA pH 8.0 (w/NaOH), 20 mM β-glycerolphosphate, complete protease inhibitors (Roche Biochemicals, Indianapolis, IN, USA) and phosphatase inhibitors (1 μg/ml Aprotinin, 1 μg/ml Leupeptin, 1 mM AEBSF, 1 mM Na3VO4, 5 mM NaF and 2.5 mM Microcystin-LR)). Each sample (10 mg proteins per well) was loaded and separated based on the molecular weights by SDS-PAGE and electrotransferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% nonfat dry milk with 0.1% Tween-20 in PBS (PBST) for 1 h at room temperature and incubated with primary antibodies at 4 °C for overnight. The primary antibodies used were mouse monoclonal anti-GFAP antibody (1 : 3000; Sigma), rabbit polyclonal anti-Iba1 antibody (1 : 500; Wako Chemicals, Richmond, VA, USA), rabbit polyclonal anti-MBP antibody (1 : 1000; Abcam, Cambridge, MA, USA), rabbit polyclonal anti-SOD2 antibody (1 : 2000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal anti-HO-1 antibody (1 : 5000; Stressgen Bioreagents, San Diego, CA, USA), mouse monoclonal anti-mitofilin antibody (1 : 500; Mitosciences, Eugene, OR, USA), rabbit polyclonal anti-PGC-1α (1 : 1000; Santa Cruz Biotechnology), mouse monoclonal anti-total OXPHOS Cx antibody (containing a mixture of antibodies to CxI-IV and ATP synthase, 1 : 3000; Invitrogen), rabbit polyclonal anti-porin antibody (1 : 2000; Calbiochem, Rockland, MA, USA) and mouse monoclonal anti-actin antibody (1 : 5000; Millipore, Billerica, MA, USA). After several washes with PBST, the membranes were incubated for 1 h at room temperature with horseradish peroxidase-conjugated goat anti-mouse IgG (1 : 5000; Bio-Rad, Hercules, CA, USA) or goat anti-rabbit IgG (1 : 5000; Bio-Rad) and developed by chemiluminescence detection. The density of protein bands from the scanned images was analyzed using ImageJ (http://www.rsb.info.nih.gov/ij/).
Immunocytochemical analysis
Immunofluorescent staining of cultured ONH astrocytes was performed as previously described.11 Briefly, the cells were fixed with 4% paraformaldehyde (Sigma)/PBS for 1 h at room temperature. To prevent nonspecific background, the cells were incubated in 1% BSA/PBS for 1 h at room temperature before incubation with mouse monoclonal anti-GFAP antibody (1 : 300; Sigma) and rabbit polyclonal anti-SOD2 antibody (1 : 2000; Santa Cruz Biotechnology) for 16 h at 4 °C. After several wash steps, the cells were incubated with the secondary antibody, Alexa Fluor 488 dye-conjugated goat anti-mouse IgG antibody (1 : 100; Invitrogen) or Alexa Fluor 488 dye-conjugated goat anti-rabbit IgG antibody (1 : 100; Invitrogen) for 4 h at 4 °C and subsequently washed with PBS. The cells were counterstained with the nucleic acid stain Hoechst 33342 (1 μg/ml, Life Technologies, Grand Island, NY, USA) in PBS. Images were acquired with confocal microscopy (Olympus FluoView1000; Olympus, Tokyo, Japan).
Morphology analysis for mitochondria
Mitochondria in the ONH astrocytes were labeled by the addition of a red fluorescent mitochondrial dye to the cultures (100 nM final concentration; MitoTracker Red CMXRos; Invitrogen-Molecular Probes) and maintaining it for 20 min in a CO2 incubator. This dye is concentrated in active mitochondria by a process that is dependent on mitochondrial membrane potential (i.e., accumulation is inhibited by actinomycin A but not by rotenone). The cultures were subsequently fixed with 4% paraformaldehyde (Sigma) in DPBS for 30 min at 4 °C and counterstained with Hoechst 33342 (1 μg/ml; Invitrogen-Molecular Probes) in DPBS. For a 3D reconstruction, images were obtained with optical section separation (z-interval) of 0.25 μm by Olympus FluoView1000. Isosurface rendition was obtained from the stack using Imaris 6.4.2 (Bitplane AG, Zurich, Switzerland).
Electron microscopy
For conventional electron microscopy, cultured ONH astrocytes after treatment were fixed with 2% paraformaldehyde, 2.5% glutaraldehyde (Ted Pella, Redding, CA, USA) in 0.15 M sodium cacodylate (pH 7.4) at 37 °C and placed in pre-cooled fixative on ice for 1 h. The following procedure was used to optimize mitochondrial structural preservation and membrane contrast. The cells were post-fixed with 1% osmium tetroxide, 0.8% potassium ferrocyanide, 3 mM calcium chloride in 0.1 M sodium cacodylate (pH 7.4) for 1 h, washed with ice-cold distilled water, post-stained with 2% uranyl acetate at 4 °C, dehydrated using graded ethanols, and embedded in Durcupan resin (Fluka, St. Louis, MO, USA). Ultrathin (70 nm) sections were post-stained with uranyl acetate and lead salts and evaluated by a JEOL 1200FX (JEOL, Tokyo, Japan) TEM operated at 80 kV. Images were recorded on film at × 8000 magnification. The negatives were digitized at 1800 dpi using a Nikon Cool scan system (Nikon Instruments Inc., Melville, NY, USA), giving an image size of 4033 × 6010 pixel array and a pixel resolution of 1.77 nm. Mitochondrial lengths were measured with ImageJ. The mitochondrial volume density, defined as the volume occupied by mitochondria divided by the volume occupied by the cytoplasm, was estimated using stereology as follows. A 112 × 112 square grid (112 × 112 chosen for ease of use with Photoshop) was overlaid on each image loaded in Photoshop (Adobe Systems Inc., San Jose, CA, USA), and the mitochondria and cytoplasm lying under intercepts were counted. The relative volume of the mitochondria was expressed as the ratio of intercepts coinciding with this organelle relative to the intercepts coinciding with the cytoplasm.
Electron microscope tomography
Sections of cells from each group were cut at thicknesses of 400–500 nm. Sections were then stained 30 min in 2% aqueous uranyl acetate, followed by 15 min in lead salts. Fiducial cues consisting of 20 nm and colloidal gold particles were deposited on opposite sides of the section. For each reconstruction, a series of images at regular tilt increments was collected with a JEOL 4000EX intermediate-voltage electron microscope operated at 400 kV. The specimens were irradiated before initiating a tilt series in order to limit anisotropic specimen thinning during image collection. Tilt series were recorded using a 4 × 4 k2 CCD camera with an angular increment of 2° from −60° to +60° about an axis perpendicular to the optical axis of the microscope using a computer-controlled goniometer to increment accurately the angular steps. The illumination was held to near parallel beam conditions and optical density was maintained constant by varying the exposure time. The magnification was 12 000 and the pixel resolution was 1.2 nm. The IMOD package (http://bio3d.colorado.edu/imod/) was used for rough alignment with the fine alignment and reconstruction was performed using the TxBR package (National Center for Microscopy and Imaging Research, San Diego, CA, USA). Volume segmentation was performed by manual tracing in the planes of highest resolution with the program Xvoxtrace.58 The mitochondrial reconstructions were visualized using Analyze (Mayo Foundation, Rochester, MN, USA) or the surface-rendering graphics of Synu (National Center for Microscopy and Imaging Research, San Diego, CA, USA) as described.58 These programs allow one to step through slices of the reconstruction in any orientation and to track or model features of interest in three dimensions.
Cell viability measurement
Cell viability was measured using 3-[4,5-dimethylthiazol-2yl]-2,5-diphenyl tetrazolium bromide (MTT) according to the manufacturer’s recommendations (Cell Proliferation Kit 1; Roche Diagnostics, Indianapolis, IN, USA). Briefly, ONH astrocytes were plated on a 96-well plate (1 × 103 per well) and after 24 h, the cells were pre-incubated with 50 μg/ml CoQ10 for 24 h, and then exposed to H2O2. At various times, a mixture of 100-ml fresh medium and 10-μl MTT stock solution was added to each well including the negative control. The cells were incubated for 4 h in a humidified atmosphere of a 5% CO2 incubator at 37 °C, 100 μl of solubilization solution was added per well to resolve the crystallized formazan. After incubation for 16–18 h in a humidified atmosphere of a 5% CO2 incubator at 37 °C, the absorbance at 560 nm was measured with a microplate reader (Spectra MAX; Molecular Devices Corp., Sunnyvale, CA, USA). Each set of data was collected from multiple replicate wells of each experimental group (n=3).
ROS measurement
The intracellular ROS was measured by 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA, Life Technologies), a chloromethyl derivative of H2DCFDA, useful as an indicator for ROS in cells. Briefly, ONH astrocytes were plated on a six-well plate (1.7 × 104 per well) and after 24 h, cells were pre-incubated with 50 μg/ml CoQ10 for 24 h, and then exposed to H2O2. The cells were detached with typsin/EDTA and loaded with 20 μM CM-H2DCFDA at 37 °C for 20 min, and then fluorescence of the sample was measured immediately using flow cytometry (BD FACSCanto II, BD Bioscience, San Diego, CA, USA). Each set of data was collected from multiple replicate dishes of each experimental group (n=3).
Cellular ATP measurement
ATP levels were measured using a luciferase-based assay kit (CellTiter-Glo, Promega Corp., Madison, WI, USA). Briefly, ONH astrocytes were plated on a 96-well plate (1 × 103 per well) and after 24 h, the cells were pre-incubated with 50 μg/ml CoQ10 for 24 h, and then exposed to H2O2. ATP standard curves from 0 to 1000 nM of ATP in growth media were prepared. After H2O2 treatment, 100 μl growth media was added, followed by 100 μl of the CellTiter-Glo reagent per well to lyse cells using vigorous shaking for 2 min at room temperature. At this point, the prepared ATP standard solution was aded. To stabilize the luminescence signal, cells at room temperature were incubated for 10 min. The luminescence was measured using a microplate luminometer (Labsystems Luminoskan, GMI Inc., Minneapolis, MN, USA). Each set of data was collected from multiple replicate wells of each experimental group (n=3).
Statistical analysis
Data were presented as the mean±S.D. or S.E.M. Comparison of two or three experimental conditions was evaluated using the unpaired, two-tailed Student’s t-test or one-way analysis of variance and the Bonferroni t-test. P<0.05 was considered statistically significant.
Abbreviations
- ATP:
-
adeonsine triphosphate
- BSA:
-
bovine serum albumin
- CoQ10:
-
coenzyme Q10
- CM-H2DCFDA:
-
5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester
- Cx:
-
complex
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- DPBS:
-
Dulbecco’s phosphate-buffered saline
- FBS:
-
fetal bovine serum
- GFAP:
-
glial fibrillary acidic protein
- H2O2:
-
hydrogen peroxide
- HO-1:
-
heme oxygenase-1
- IOP:
-
intraocular pressure
- MBP:
-
myelin basic protein
- MTT:
-
3-[4,5-dimethylthiazol-2yl]-2,5-diphenyl tetrazolium bromide
- ONH:
-
optic nerve head
- OXPHOS:
-
oxidative phoshorylation
- PBST:
-
0.1% Tween-20 in PBS
- PGC-1α:
-
peroxisome-proliferator-activated receptor-γ coactivator-1
- RGC:
-
retinal ganglion cell
- ROS:
-
reactive oxygen species
- SA:
-
surface area
- SOD2:
-
superoxide dismutase 2
References
Cadenas E, Davies KJ . Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med 2000; 29: 222–230.
Chrysostomou V, Rezania F, Trounce IA, Crowston JG . Oxidative stress and mitochondrial dysfunction in glaucoma. Curr Opin Pharmacol 2013; 13: 12–15.
McElnea EM, Quill B, Docherty NG, Irnaten M, Siah WF, Clark AF et al. Oxidative stress, mitochondrial dysfunction and calcium overload in human lamina cribrosa cells from glaucoma donors. Mol Vis 2011; 17: 1182–1191.
Abu-Amero KK, Morales J, Bosley TM . Mitochondrial abnormalities in patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci 2006; 47: 2533–2541.
Lee D, Kim KY, Noh YH, Chai S, Lindsey JD, Ellisman MH et al. Brimonidine blocks glutamate excitotoxicity-induced oxidative stress and preserves mitochondrial transcription factor a in ischemic retinal injury. PLoS One 2012; 7: e47098.
Hernandez MR, Agapova OA, Yang P, Salvador-Silva M, Ricard CS, Aoi S . Differential gene expression in astrocytes from human normal and glaucomatous optic nerve head analyzed by cDNA microarray. Glia 2002; 38: 45–64.
Weinreb RN, Khaw PT . Primary open-angle glaucoma. Lancet 2004; 363: 1711–1720.
Hernandez MR . The optic nerve head in glaucoma: role of astrocytes in tissue remodeling. Prog Retin Eye Res 2000; 19: 297–321.
Tezel G . Oxidative stress in glaucomatous neurodegeneration: mechanisms and consequences. Prog Retin Eye Res 2006; 25: 490–513.
Dai C, Khaw PT, Yin ZQ, Li D, Raisman G, Li Y . Structural basis of glaucoma: the fortified astrocytes of the optic nerve head are the target of raised intraocular pressure. Glia 2012; 60: 13–28.
Ju WK, Kim KY, Lindsey JD, Angert M, Duong-Polk KX, Scott RT et al. Intraocular pressure elevation induces mitochondrial fission and triggers OPA1 release in glaucomatous optic nerve. Invest Ophthalmol Vis Sci 2008; 49: 4903–4911.
Ju WK, Kim KY, Angert M, Duong-Polk KX, Lindsey JD, Ellisman MH et al. Memantine blocks mitochondrial OPA1 and cytochrome c release and subsequent apoptotic cell death in glaucomatous retina. Invest Ophthalmol Vis Sci 2009; 50: 707–716.
Ju WK, Kim KY, Duong-Polk KX, Lindsey JD, Ellisman MH, Weinreb RN . Increased optic atrophy type 1 expression protects retinal ganglion cells in a mouse model of glaucoma. Mol Vis 2010; 16: 1331–1342.
Son JL, Soto I, Oglesby E, Lopez-Roca T, Pease ME, Quigley HA et al. Glaucomatous optic nerve injury involves early astrocyte reactivity and late oligodendrocyte loss. Glia 2010; 58: 780–789.
Beal MF, Shults CW . Effects of Coenzyme Q10 in Huntington's disease and early Parkinson's disease. Biofactors 2003; 18: 153–161.
McCarthy S, Somayajulu M, Sikorska M, Borowy-Borowski H, Pandey S . Paraquat induces oxidative stress and neuronal cell death; neuroprotection by water-soluble coenzyme Q10. Toxicol Appl Pharmacol 2004; 201: 21–31.
Bessero AC, Clarke PG . Neuroprotection for optic nerve disorders. Curr Opin Neurol 2010; 23: 10–15.
Qu J, Kaufman Y, Washington I . Coenzyme Q10 in the human retina. Invest Ophthalmol Vis Sci 2009; 50: 1814–1818.
Nucci C, Tartaglione R, Cerulli A, Mancino R, Spano A, Cavaliere F et al. Retinal damage caused by high intraocular pressure-induced transient ischemia is prevented by coenzyme Q10 in rat. Int Rev Neurobiol 2007; 82: 397–406.
Russo R, Cavaliere F, Rombola L, Gliozzi M, Cerulli A, Nucci C et al. Rational basis for the development of coenzyme Q10 as a neurotherapeutic agent for retinal protection. Prog Brain Res 2008; 173: 575–582.
Nakajima Y, Inokuchi Y, Nishi M, Shimazawa M, Otsubo K, Hara H . Coenzyme Q10 protects retinal cells against oxidative stress in vitro and in vivo. Brain Res 2008; 1226: 226–233.
Lulli M, Witort E, Papucci L, Torre E, Schipani C, Bergamini C et al. Coenzyme Q10 instilled as eye drops on the cornea reaches the retina and protects retinal layers from apoptosis in a mouse model of kainate-induced retinal damage. Invest Ophthalmol Vis Sci 2012; 53: 8295–8302.
Hernandez MR, Igoe F, Neufeld AH . Cell culture of the human lamina cribrosa. Invest Ophthalmol Vis Sci 1988; 29: 78–89.
Yu AL, Fuchshofer R, Birke M, Kampik A, Bloemendal H, Welge-Lussen U . Oxidative stress and TGF-beta2 increase heat shock protein 27 expression in human optic nerve head astrocytes. Invest Ophthalmol Vis Sci 2008; 49: 5403–5411.
Hernandez MR, Miao H, Lukas T . Astrocytes in glaucomatous optic neuropathy. Prog Brain Res 2008; 173: 353–373.
John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM et al. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell 2005; 16: 1543–1554.
Van Laar VS, Dukes AA, Cascio M, Hastings TG . Proteomic analysis of rat brain mitochondria following exposure to dopamine quinone: implications for Parkinson disease. Neurobiol Dis 2008; 29: 477–489.
Marriage BJ, Clandinin MT, Macdonald IM, Glerum DM . Cofactor treatment improves ATP synthetic capacity in patients with oxidative phosphorylation disorders. Mol Genet Metab 2004; 81: 263–272.
Haas RH . The evidence basis for coenzyme Q therapy in oxidative phosphorylation disease. Mitochondrion 2007; 7 (Suppl): S136–S145.
Fragaki K, Cano A, Benoist JF, Rigal O, Chaussenot A, Rouzier C et al. Fatal heart failure associated with CoQ10 and multiple OXPHOS deficiency in a child with propionic acidemia. Mitochondrion 2011; 11: 533–536.
He Y, Leung KW, Zhang YH, Duan S, Zhong XF, Jiang RZ et al. Mitochondrial complex I defect induces ROS release and degeneration in trabecular meshwork cells of POAG patients: protection by antioxidants. Invest Ophthalmol Vis Sci 2008; 49: 1447–1458.
Malone PE, Hernandez MR . 4-Hydroxynonenal, a product of oxidative stress, leads to an antioxidant response in optic nerve head astrocytes. Exp Eye Res 2007; 84: 444–454.
Mozaffarieh M, Flammer J . Is there more to glaucoma treatment than lowering IOP? Surv Ophthalmol 2007; 52 (Suppl 2): S174–S179.
Mozaffarieh M, Flammer J . A novel perspective on natural therapeutic approaches in glaucoma therapy. Expert Opin Emerg Drugs 2007; 12: 195–198.
Mozaffarieh M, Grieshaber MC, Orgul S, Flammer J . The potential value of natural antioxidative treatment in glaucoma. Surv Ophthalmol 2008; 53: 479–505.
Lee S, Sheck L, Crowston JG, Van Bergen NJ, O'Neill EC, O'Hare F et al. Impaired complex-I-linked respiration and ATP synthesis in primary open-angle glaucoma patient lymphoblasts. Invest Ophthalmol Vis Sci 2012; 53: 2431–2437.
Yu-Wai-Man P, Griffiths PG, Chinnery PF . Mitochondrial optic neuropathies—disease mechanisms and therapeutic strategies. Prog Retin Eye Res 2011; 30: 81–114.
Yu AL, Moriniere J, Birke M, Neumann C, Fuchshofer R, Kampik A et al. Reactivation of optic nerve head astrocytes by TGF-beta2 and H2O2 is accompanied by increased Hsp32 and Hsp47 expression. Invest Ophthalmol Vis Sci 2009; 50: 1707–1717.
Shults CW, Haas RH, Passov D, Beal MF . Coenzyme Q10 levels correlate with the activities of complexes I and II/III in mitochondria from parkinsonian and nonparkinsonian subjects. Ann Neurol 1997; 42: 261–264.
Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM et al. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J Neurosci 2002; 22: 1592–1599.
Guo L, Cordeiro MF . Assessment of neuroprotection in the retina with DARC. Prog Brain Res 2008; 173: 437–450.
Lulli M, Witort E, Papucci L, Torre E, Schiavone N, Dal Monte M et al. Coenzyme Q10 protects retinal cells from apoptosis induced by radiation in vitro and in vivo. J Radiat Res 2012; 53: 695–703.
Rohl C, Armbrust E, Kolbe K, Lucius R, Maser E, Venz S et al. Activated microglia modulate astroglial enzymes involved in oxidative and inflammatory stress and increase the resistance of astrocytes to oxidative stress in vitro. Glia 2008; 56: 1114–1126.
Bruce-Keller AJ, Geddes JW, Knapp PE, McFall RW, Keller JN, Holtsberg FW et al. Anti-death properties of TNF against metabolic poisoning: mitochondrial stabilization by MnSOD. J Neuroimmunol 1999; 93: 53–71.
Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999; 98: 115–124.
Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 2002; 418: 797–801.
Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 1998; 18: 231–236.
Scarpulla RC . Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene 2002; 286: 81–89.
Fosslien E . Mitochondrial medicine--molecular pathology of defective oxidative phosphorylation. Ann Clin Lab Sci 2001; 31: 25–67.
Bargossi AM, Battino M, Gaddi A, Fiorella PL, Grossi G, Barozzi G et al. Exogenous CoQ10 preserves plasma ubiquinone levels in patients treated with 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors. Int J Clin Lab Res 1994; 24: 171–176.
Fukui H, Moraes CT . The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? Trends Neurosci 2008; 31: 251–256.
Balaban RS, Nemoto S, Finkel T . Mitochondria, oxidants, and aging. Cell 2005; 120: 483–495.
Carelli V, Rugolo M, Sgarbi G, Ghelli A, Zanna C, Baracca A et al. Bioenergetics shapes cellular death pathways in Leber's hereditary optic neuropathy: a model of mitochondrial neurodegeneration. Biochim Biophys Acta 2004; 1658: 172–179.
Beal MF . Aging, energy, and oxidative stress in neurodegenerative diseases. Ann Neurol 1995; 38: 357–366.
Bergamini C, Moruzzi N, Sblendido A, Lenaz G, Fato R . A water soluble CoQ10 formulation improves intracellular distribution and promotes mitochondrial respiration in cultured cells. PLoS One 2012; 7: e33712.
Quinzii CM, Lopez LC, Gilkerson RW, Dorado B, Coku J, Naini AB et al. Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J 2010; 24: 3733–3743.
Fernandez-Marcos PJ, Auwerx J . Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 2011; 93: 884S–890S.
Perkins GA, Renken CW, Song JY, Frey TG, Young SJ, Lamont S et al. Electron tomography of large, multicomponent biological structures. J Struct Biol 1997; 120: 219–227.
Acknowledgements
We thank Professor James D Lindsey for critical comments on the manuscript. This work was supported, in part, by NIH Grants EY018658 (WKJ) and NCRR P41 RR004050 and P41GM103412-24 (MHE), and an unrestricted grant from Research to Prevent Blindness (New York, NY, USA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Finazzi-Agró
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Noh, Y., Kim, KY., Shim, M. et al. Inhibition of oxidative stress by coenzyme Q10 increases mitochondrial mass and improves bioenergetic function in optic nerve head astrocytes. Cell Death Dis 4, e820 (2013). https://doi.org/10.1038/cddis.2013.341
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.341
Keywords
This article is cited by
-
The Effect of Organophosphate Exposure on Neuronal Cell Coenzyme Q10 Status
Neurochemical Research (2021)
-
Increasing Nrf2 Activity as a Treatment Approach in Neuropsychiatry
Molecular Neurobiology (2021)
-
AICAR and Decitabine Enhance the Sensitivity of K562 Cells to Imatinib by Promoting Mitochondrial Activity
Current Medical Science (2020)
-
Resveratrol Protects Optic Nerve Head Astrocytes from Oxidative Stress-Induced Cell Death by Preventing Caspase-3 Activation, Tau Dephosphorylation at Ser422 and Formation of Misfolded Protein Aggregates
Cellular and Molecular Neurobiology (2020)
-
MitoQ and CoQ10 supplementation mildly suppresses skeletal muscle mitochondrial hydrogen peroxide levels without impacting mitochondrial function in middle-aged men
European Journal of Applied Physiology (2020)