Abstract
The transcription factor p63 is required for proper epidermal barrier formation and maintenance. Herein, we used chromatin immunoprecipitation coupled with DNA sequencing to identify novel p63 target genes involved in normal human epidermal keratinocyte (NHEKs) growth and differentiation. We identified over 2000 genomic sites bound by p63, of which 82 were also transcriptionally regulated by p63 in NHEKs. Through the discovery of interleukin-1-α as a p63 target gene, we identified that p63 is a regulator of epithelial–mesenchymal crosstalk. Further, three-dimensional organotypic co-cultures revealed TCF7L1, another novel p63 target gene, as a regulator of epidermal proliferation and differentiation, providing a mechanism by which p63 maintains the proliferative potential of basal epidermal cells. The discovery of new target genes links p63 to diverse signaling pathways required for epidermal development, including regulation of paracrine signaling to proliferative potential. Further mechanistic insight into p63 regulation of epidermal cell growth and differentiation is provided by the identification of a number of novel p63 target genes in this study.
Similar content being viewed by others
Main
p63, a homolog of p53, is required for proper epidermal development.1, 2 Owing to multiple transcriptional promoters in the 5′ end of the gene, p63 is expressed either containing (TA) or lacking (ΔN) an N-terminal acidic transactivation domain. Further, alternative C-terminal processing results in one of three splice variants (α, β or γ). Although a number of isoforms are possible, ΔNp63α is the predominantly expressed isoform in human epithelial cells.3, 4, 5 ΔNp63 was originally characterized as having dominant-negative properties toward TAp63 isoforms and p53;6 however, ΔNp63 can also positively regulate transcription owing to an additional N-terminal transactivation domain.7, 8
p63−/− mice display severe developmental defects, the most notable being a complete lack of complex epithelia.1, 2 Further, these phenotypes are recapitulated in humans bearing heterozygous germline p63 mutations.9, 10 p63 expression is localized to the basal layer of complex epithelia and it has been speculated that p63 is required to maintain the proliferative potential of epidermal stem cells.1, 11 Additional studies have suggested that p63 is crucial for commitment of stem cells to an epidermal cell fate.2 Ectopic p63 expression is sufficient to induce stratification of a single-layered epithelium,12 suggesting that it is a critical regulator of epidermal differentiation.
Several approaches have been used to identify downstream effectors of p63, including microarray analysis,4, 5, 13, 14 chromatin immunoprecipitation (ChIP)-based methods,15, 16 and ChIP-on-ChIP.17, 18 Results from these studies provide valuable insight into the mechanisms by which p63 controls molecular signaling. However, the vast majority of studies aimed at identifying p63-regulated signaling pathways have used established, immortalized and tumor-derived cell lines, all of which may not be ideally suited for understanding p63 function in normal, non-tumorigenic basal epithelium. The aim of this study was to identify p63-regulated signaling pathways that function in normal human epithelial cells.
Herein, we used a ChIP-based approach coupled with gene expression analysis of primary human epidermal keratinocytes (NHEKs) to identify transcriptional targets of p63. We identified over 2000 p63-bound genes and 338 p63-regulated genes, of which 82 genes were direct p63 target genes. By analyzing the role(s) of select target genes in epidermis development, we provide mechanistic insight into signaling pathways and functions regulated by p63 during expansion and maintenance of stratified epithelia.
Results
Identification of genome-wide p63 binding sites
To identify p63-bound genomic loci in rapidly proliferating normal human basal epithelial cells, we used a ChIP-based discovery method and primary cultures of NHEKs, as they express robust levels of ΔNp63α (Figure 1a) and are able to recapitulate epidermal differentiation in a two- and three-dimensional culture system.3, 19 NHEKs express low levels of a faster migrating protein that crossreacts with the p63-specific antibody; however, it does not migrate at the same position as any p63 isoforms by SDS-PAGE and is likely a breakdown product (ref.3 and Figure 1a). ChIP was performed to purify chromatin-bound p63 using the H129 polyclonal antibody that specifically recognizes p63α isoforms. Initial analysis revealed an enrichment of known p63 binding sites located near the p21, MDM2, and Jagged-1 genes (Figure 1b) and validated our immunoprecipitation conditions.
Identification of novel direct p63 target genes. (a) Rapidly growing cultures of NHEKs were analyzed for p63 isoforms by comparison with protein markers generated by transfection of the indicated expression vectors in H1299 cells. Total protein was isolated and expression of p63 isoforms was analyzed by western blot. (b) NHEKs were chemically crosslinked and quantitative chromatin immunoprecipitation used to determine the relative binding of p63 to a negative control region (con) and to three previously published p63 binding sites located in the p21, MDM2, and Jagged-1. (c) p63-bound genes in NHEKs were overlayed with p63 target genes identified in ME180 cells.17 Bootstrap distribution statistical analysis was performed to compare the significance of gene overlap between the two datasets. (d) NHEKs were transduced with a retrovirus-expressing shRNAs specific to GFP (con) or the DNA-binding domain of p63 (sip63). Cells were puromycin selected for 48 h and harvested for western analysis of p63 and actin. (e) RNA was isolated and submitted, in duplicate, for Affymetrix microarray analysis to identify genes with expression patterns dependent on the presence of p63. Data shown are representative of at least three independent experiments. p63-bound genes identified in the ChIP dataset (left circle) were overlayed with genes exhibiting p63-dependent expression in microarray analyses (right circle). Eighty-two genes, as annotated by the GeneSpring 7 software, were present in both datasets and were chosen for further biological analyses
Having validated our pool of chromatin fragments, we used GenPathway FactorPath Discovery technology to identify novel sites bound by p63. FactorPath Discovery has previously been used to identify genomic fragments bound by a number of DNA-interacting proteins.20, 21 Over 5000 p63-bound chromatin fragments were isolated, sequenced, and mapped to the human genome, with sequences mapping within 10 kb of known genes considered for further analysis. Using these criteria resulted in 2879 p63-bound genes (see Materials and Methods). Numerous previously identified p63 target genes were present in this dataset, including p21, MDM2, stratifin, IGFBP-3, PTPN14, RUNX1, P-cadherin, S100A2, and claudin-1, among others. Moreover, comparison of our dataset with a previously published p63 ChIP-on-ChIP17 identified 874 genes (P<0.0001) common between both datasets (Figure 1c), further validating the ability of our assay to identify novel p63 genomic binding sites.
Discovery of novel p63 target genes in NHEKs
As transcription factor binding to a gene does not necessitate transcriptional regulation, we performed microarray analysis to determine which of the identified p63 binding sites were associated with transcriptional regulation of nearby genes. NHEKs were transduced with a retrovirus expressing an shRNA targeting the DNA-binding domain of p63 (p63DBD) or GFP as a control,4 puromycin selected, and harvested for protein and RNA. Significant p63 knockdown was observed at the protein level (Figure 1d). As a threshold, we analyzed all genes displaying a minimum of two-fold change in transcript levels after p63 depletion. Using these criteria, we identified 338 genes displaying a p63-dependent expression pattern (Supplementary Table S1). Of these, 82 were present in the ChIP dataset (Figure 1e) and we focused subsequent analyses on the 82 genes (Supplementary Table S2).
Functional annotation of the 82 target genes provided additional validation and insight into the role of p63 in biological processes (Supplementary Table S3). ‘Cellular movement’, one of the most significantly enriched categories represented by the p63 target genes, is supported by recent data identifying p63 as a regulator of migration and the epithelial-to-mesenchymal transition.4, 22, 23 In addition, a number of functional annotations are supported by the phenotype of p63−/− animals, such as ‘cellular development’, ‘tissue development’, and ‘hair and skin development and function’.1, 2 Overall, associating the 82 candidate genes with enriched functional processes and pathways added further confidence that our approach identified valid p63 target genes.
As a secondary screen, we performed semiquantitative ChIP using a unique p63 antibody (4A4) than used to generate the initial ChIP dataset (H129). For this validation, we focused on a subset of 14 genes exhibiting a minimum of two-fold regulation by p63, and that we could link to epidermal biology through previous publications. The known p63 binding site located in the p21 promoter and an exon sequence located in the GAPDH gene were used as positive and negative controls, respectively. Statistically significant p63 binding to promoter or intronic sequences was observed for 11 of the 14 genes analyzed (Figure 2).
Validation of p63-bound response elements located in/near target genes. Rapidly growing NHEKs were chemically crosslinked, snap frozen, and submitted for FactorPath query analysis. Specific p63 binding sites were selected from the regions identified by the initial ChIP experiment. A majority of the sites queried were enriched by p63 immunoprecipitation, with statistically significant hits marked with an asterisk (*P<0.05; **P<0.01). Data presented were generated from three independent experiments. Please note that for BDKRB2, we identified two independent p63 binding sites, denoted (1) and (2)
In secondary analyses to confirm p63-dependent transcriptional regulation of select target genes, NHEKs were transduced with siRNA-expressing adenovirus targeting the p63DBD, or shuttle adenovirus as a control.5 This knockdown strategy is unique from the retrovirus used to generate initial microarray datasets. After successful p63 depletion at the protein level (Figure 3a), qPCR analyses revealed that all target genes analyzed displayed p63-regulated transcript changes that were consistent with those observed in our initial microarray observations (Figure 3b), validating these genes as bona fide p63-regulated genes.
Validation of genes displaying a p63-dependent expression pattern. (a) NHEKs were transduced with adenovirus-expressing p63DBD siRNA, or shuttle virus as a control. Protein lysates were harvested 48 h later and western blot analysis performed for p63 and actin. (b) Rapidly growing NHEKs were treated as in (a) and RNA was isolated 48 h after infection and target genes analyzed by quantitative real-time PCR. Data presented are representative of three independent experiments. (c) Three TAp63-specific siRNAs were co-transfected with TAp63γ into H1299 cells. The expression of TAp63 was analyzed as in (a). (d) TAp63 siRNAs were co-transfected with TAp73β. p73-specific siRNAs were used as a positive control for knockdown. Expression of TAp73 and actin were analyzed as in (a). (e) TAp63 siRNAs were transfected into NHEKs and levels of ΔNp63α, p53, and actin were analyzed by western blot. (f) NHEKs were transfected with TAp63-specific siRNAs and qPCR was used to analyze endogenous levels of ΔNp63 and TAp63 transcript levels 48 h after transfection. (g) NHEKs were transfected with NS, or TAp63-specific siRNAs as in (b), and RNA was analyzed by quantitative real-time PCR to observe transcript levels of a subset of novel direct p63 target genes. Each of the three siTAp63 oligos were transfected into independent cultures of NHEKs, and data presented next to the original microarray data (gray bar) represent the mean of all three TAp63 siRNA experiments (black bar)
Despite its low, often undetectable protein levels, recent evidence suggests that endogenous TAp63 isoforms can regulate target gene expression and differentiation of primary epithelial cells.19, 24 As our siRNAs target all p63 isoforms, we designed independent siRNAs that specifically target TAp63 and tested the efficacy of each against exogenous TAp63γ in H1299 cells, which lack detectable expression of any p53 family members. We observed a robust knockdown of TAp63γ at the protein level (Figure 3c). Further, we confirmed that these siRNAs do not target TAp73β (Figure 3d), or endogenous p53 and ΔNp63 isoforms (Figure 3e). To test the dependency of target gene expression on TAp63, we transfected non-silencing (NS) or TAp63-specific siRNAs into NHEKs to deplete endogenous TAp63 levels. Specific knockdown of endogenous TAp63, but not ΔNp63, isoforms was confirmed by qPCR (Figure 3f). At 48λh after transfection, target gene expression was determined and compared with those changes observed after depleting all p63 isoforms (compare 3G to 3B). Although all analyzed genes were dependent on ΔNp63 isoforms for expression, consistent with ΔNp63α being the predominant isoform expressed in epithelial cells, few, if any, genes showed a significant dependence on TAp63 for expression. Taken collectively, these data suggest that, of the target genes analyzed, ΔNp63 isoforms function as the primary regulator of transcriptional regulation.
p63 target genes correlate with p63 expression in squamous cell carcinoma
Consistent with the role of p63 in maintaining proliferative potential in normal basal epithelium,1 amplification of its genomic locus is observed in squamous cell carcinomas,25 particularly those derived from tissues of the head and neck (HNSCC). To analyze the expression of our novel p63 target genes versus p63 in an in vivo setting, we used two publicly available HNSCC gene expression datasets.26, 27 We used the gene expression profiles of tumors exhibiting high versus low p63 expression (Supplementary Figure S1A) and compared the expression of our target genes between each subset. Of the 82 novel p63 targets, 25 genes exhibited differential expression patterns in HNSCC, when clustered for p63 expression (Supplementary Figure S1B). Of note, many p63 target genes were not differentially expressed in these datasets. We speculate that the latter is owing to differential expression of p53, p73, or other co-factors between normal and tumor cells, or the relative contribution of TAp63 versus ΔNp63 expression, which was not possible to assess from the tumor microarray expression data. Nevertheless, the correlation between 25 target genes and p63 status in an in vivo setting further supports our findings that these target genes contribute to p63-mediated biological processes in both normal and tumorigenic settings. Given this observation, we chose to functionally analyze two of these genes and determine the mechanism by which they contribute to p63-regulated biological processes.
p63 regulates paracrine signaling between epidermal keratinocytes and dermal fibroblasts
Interleukin-1-α (IL1A) is a secreted factor important for maintaining epidermal homeostasis by regulating fibroblast-derived cytokines.28 Expressed in basal keratinocytes, IL1A is secreted to dermal fibroblasts where it induces the expression of fibroblast-derived growth factors, such as KGF, GM-CSF, and FGF10.29 These factors, in return, induce both the proliferation and differentiation of epidermal keratinocytes, thus completing an essential paracrine loop (Figure 4a).30, 31
p63 regulates paracrine pathways between keratinocytes and dermal fibroblasts. (a) Model of paracrine signaling between the epidermis and the dermis. (b) NHEKs were transiently transfected with control (NS) or p63-specific siRNA, harvested 72 h later, and p63 and actin protein levels analyzed by western blot. (c) Conditioned medium from transfected NHEKs was collected 72 h after transfection and secreted IL1A concentration was determined by ELISA. (d) NHEK-conditioned medium (cm) was obtained by growing NHEKs for 48 h, filtered, and given as the only media to NHDFs for 24 h. Unconditioned NHEK medium (ucm) was used as a control. NHDFs were harvested 24 h after the addition of medium and harvested for RNA. Quantitative real-time PCR was used to quantify the expression of the fibroblast-specific growth factors KGF and FGF10 in response to NHEK-conditioned medium, relative to an unconditioned control. (e) NHDFs were treated as in (d) and GM-CSF levels were analyzed and graphed independently owing to the robust induction observed following cm addition. (f) NHEKs were transfected with NS or IL1A-specific siRNA and IL1A depletion was confirmed by qPCR 48 h after transfection. (g) NHEKs were transfected with p63 or IL1A siRNA alone or in combination, as well as non-silencing siRNA (NS) as a control. At 48 h after transfection, the media were replaced, conditioned medium was generated, and NHDFs were treated as in (d). KGF expression was measured by qPCR 24 h after the addition of conditioned medium. (h) NHDFs treated as in (g) and qPCR was used to analyze the expression of GM-CSF or FGF10 (i). (j) NHEKs were transduced with shuttle (sicon) or p63 siRNA- (sip63) expressing adenovirus. Cells were harvested, RNA isolated, and quantitative real-time PCR performed to analyze the expression of p63 and KGFR. All quantitative PCR data were normalized to actin levels and error bars represent standard deviation of at least three independent experiments. *P<0.5
The identification of IL1A as a p63 target gene in keratinocytes led us to hypothesize that p63 regulation of secreted cytokines is important for paracrine control of fibroblast-derived cytokines that are essential for proper development and maintenance of the epidermis. Consistent with this hypothesis, siRNA-mediated depletion of p63 in NHEKs (Figure 4b) resulted in significantly lower levels of secreted IL1A, as measured by ELISA (Figure 4c). To determine whether NHEKs could induce fibroblast-derived cytokine expression in a paracrine manner, we cultured dermal fibroblasts (NHDFs) in media conditioned by NHEKs. After NHDF growth in NHEK-conditioned medium for 48 h, we observed a significant induction of the fibroblast-derived growth factors, KGF, FGF10, and GM-CSF, relative to unconditioned keratinocyte media (Figures 4d and e). To test our hypothesis that p63 regulates, in a paracrine manner, the expression of fibroblast-specific cytokines, we transfected NHEKs with control, p63, or IL1A-specific siRNA, and collected conditioned media from the cultures. IL1A depletion was verified by qPCR analysis in NHEKs (Figure 4f). NHDFs cultured in the medium conditioned by NHEKs depleted of p63 had significantly reduced the production of KGF, FGF10, and GM-CSF, relative to control cells (Figures 4g–i), suggesting that p63 signaling in keratinocytes is important for promoting epidermis–dermis paracrine signaling. Of note, these data were not a result of overall decrease in NHEK cell number following p63 knockdown, as we observed minimal cell death and detachment following transfection (data not shown). Interestingly, we observed a similar, slightly less robust, inhibition of cytokine production using the conditioned medium from NHEKs expressing reduced levels of IL1A (Figures 4g–i). We were not able to further inhibit cytokine production by depleting p63 and IL1A concomitantly, suggesting that the majority of paracrine signaling controlled by p63 is owing to its regulation of IL1A expression.
Of these fibroblast-derived cytokines, KGF binds to its cognate keratinocyte receptor, KGFR, on basal epidermal cell, and it acts as a potent mitogen to induce proliferation of epidermal progenitor cells. We hypothesized that p63 may also regulate the expression of KGFR in basal epidermal cells. Indeed, the expression of KGFR was significantly reduced after p63 knockdown (Figure 4j), suggesting that p63 acts to regulate KGFR expression in basal keratinocytes. Collectively, through the identification of IL1A as a p63 target gene, we discovered a role for p63 in regulating paracrine signaling pathways between epidermal keratinocytes and dermal fibroblasts.
TCF7L1 regulates a proliferation–differentiation switch in three-dimensional organotypic cultures
TCF7L1 is a downstream component of the Wnt signaling pathway. It acts as an effector to control the transcriptional state of Wnt-regulated target genes. Recent data suggest that TCF7L1 can control cell fate lineage commitment in multipotent skin stem cells.32 In addition, induction of TCF7L1 in mouse epidermal progenitor cells is sufficient to abrogate terminal differentiation and maintain an undifferentiated cell fate.33
We analyzed TCF7L1 expression during calcium-induced NHEK differentiation in monolayer. Consistent with p63 expression, TCF7L1 levels correlated with the undifferentiated state of keratinocytes and significantly decreased as cells underwent terminal differentiation (Figures 5a and c, control lanes). From these data, we hypothesized that TCF7L1 is important for maintaining the undifferentiated state of p63-expressing, rapidly growing NHEKs. To test this hypothesis, we used control or TCF7L1-overexpressing NHEKs and induced terminal differentiation in culture. Ectopic TCF7L1 expression decreased mRNA levels of filaggrin, loricrin, and involucrin (Figure 5b), substantially reduced the protein levels of loricrin and crosslinked involucrin (Figure 5c), and thus attenuated differentiation. In both settings, calcium-induced differentiation decreased ΔNp63α levels by 6 days, providing additional evidence that TCF7L1 functions downstream of p63 (Figure 5c). These data further supported our hypothesis that TCF7L1 is crucial for maintaining a rapidly growing, undifferentiated state of p63-expressing keratinocytes.
TCF7L1 is a negative regulator of keratinocyte differentiation. (a) At confluency, rapidly growing monolayers of NHEKs were treated with 1.4 mM calcium to induce differentiation. Cultures were harvested at the indicated time points, RNA isolated, and qRT-PCR performed to determine the expression of p63 and TCF7L1 during differentiation. Expression levels were normalized to actin levels and presented relative to cultures at day 0. Error bars represent standard deviation of three replicate experiments. (b) NHEKs were transduced with control or ectopic TCF7L1-expressing lentivirus and induced to differentiate 48 h after infection. Cultures were collected at indicated time points and RNA was isolated for qPCR analysis of differentiation genes. Error bars represent standard deviation of three independent experiments. (c) NHEKs were treated as in (b), protein harvested at indicated time points, and levels of TCF7L1, p63, involucrin, loricrin, and actin analyzed by western blot
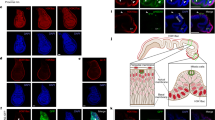
To determine the function of endogenous TCF7L1 in NHEKs, we used a three-dimensional organotypic co-culture (OTC) model system to analyze differentiation in control NHEKs as compared with TCF7L1-deficient NHEKs. Like cultured NHEKs grown in the monolayer, the predominant isoform of p63 expressed in epidermal portion of OTCs is ΔNp63α (Supplementary Figure S2). To modulate endogenous TCF7L1 levels, we transfected NHEKs with TCF7L1-specific siRNAs, as well as with NS siRNAs to serve as a control. Although significant TCF7L1 depletion was achieved, endogenous p63 protein levels were not altered (Figure 6a). At 24 h after transfection, NHEKs were plated onto gels composed of rat-tail collagen embedded with normal human dermal fibroblasts and lifted to the air–liquid interface. After 10 days, cultures were harvested and in situ expression of various proteins analyzed by immunofluorescence. In control cultures, involucrin expression was detected in the upper spinous layers, whereas filaggrin, loricrin, and keratin 2 were largely restricted to the stratum granulosum (Figure 6b). In TCF7L1-depleted cultures, we observed an earlier onset of all observed differentiation markers, with involucrin being present throughout all suprabasal layers. Furthermore, filaggrin, loricrin, and keratin 2, typically restricted to the stratum granulosum, were already detectable in spinous layers of the epidermis (Figure 6b). We did not observe measurable differences in the cell–cell contact protein E-cadherin, suggesting that the effects of TCF7L1 depletion is limited to proteins involved in terminal differentiation. These data suggest that keratinocytes, when depleted of TCF7L1 expression, prematurely initiate a program of terminal differentiation in OTC.
Loss of TCF7L1 results in early differentiation of NHEKs in OTC. (a) NHEKs were transfected with siRNAs targeting TCF7L1 or non-silencing oligos (NS) as a control. At 48 h after transfection, RNA was isolated and qRT-PCR performed to assess the expression of TCF7L1. (b) Immunofluorescent analysis of control and siTCF7L1 OTCs were performed with antibodies specific for filaggrin, involucrin, loricrin, keratin 2, and E-cadherin. Dotted line represents the epidermis–dermis interface. For proliferation analysis, OTCs were grown in medium containing BrdU for 16 h before harvesting in order to label proliferative cells. Immunofluorescence with a BrdU-specific antibody was performed to identify actively proliferating cells in the epidermal layers of OTCs. (c) BrdU-positive cells from (b) were quantified and graphed. Data shown are representative of three independent experiments
We hypothesized that following TCF7L1 depletion, increased differentiation would be accompanied by decreased proliferation. As seen in Figures 6b and c, there was a significant decrease in the number of 5-bromodeoxyuridine (BrdU)-positive cells in siTCF7L1 cultures compared with control cultures. Coupled with our ectopic TCF7L1 data, our studies suggest that TCF7L1-depleted keratinocytes are unable to maintain an undifferentiated state. Moreover, we identified a number of Wnt target genes that displayed significant expression changes following p63 depletion in NHEKs (Supplementary Figure S3), consistent with the transcriptional regulatory function of TCF7L1. Our combined use of a genomic approach to identify new target genes, and an organotypic assay to screen for functions downstream of p63, allowed us to mechanistically link p63 signaling to a critical proliferation/differentiation switch (TCF7L1) in stratified epithelia.
Further identification of novel p63 target genes in NHEKs
A recent study by Truong et al.19 reported that 231 genes were regulated in a p63-dependent manner in NHEKs grown in an organotypic culture. However, the experimental design did not reveal which of these genes were direct targets of p63 regulation. By overlaying our ChIP dataset with the microarray dataset from this previous study, we identified 32 additional genes as directly regulated by p63 in primary keratinocytes (Table 1). Thus, we were able to extend the use of our ChIP dataset outside this study to further identify novel p63 target genes using publicly available expression datasets.
Discussion
In this study, we used a ChIP-based methodology, coupled with expression microarray, to identify p63 binding sites in NHEKs. Although previous studies have used ChIP-based approaches to identify p63-bound genomic regions in numerous epithelial models,16 ours is the first study in primary human keratinocytes. We discovered 874 genes (P<0.0001) in common between our ChIP dataset and a previously published ChIP-on-ChIP dataset.17 Despite significant overlap, the large number of genes unique to p63 regulation in NHEKs, versus the transformed ME180 cell line, suggests that there are significant differences between p63 signaling in normal keratinocytes as compared with immortalized and/or tumorigenic settings.
Coupling ChIP-based data with microarray analyses allowed for rapid identification of 82 p63 target genes in NHEKs, and secondary validation of our datasets were crucial in ruling out false-positive genes. This validation included the use of a second p63 antibody to confirm the ChIP results, along with the use of a unique p63-specific siRNA to corroborate the microarray expression dataset. Furthermore, we identified in vivo correlations between the expression of p63 and direct target genes in HNSCC, suggesting that these genes are important for mediating the function of p63 in a tumorigenic environment. We are currently working to identify the role(s) of these genes in a tumorigenic setting, and these studies will provide insight into how p63 functions in both normal and tumor-derived epithelia.
Expressed in basal, proliferating, keratinocytes, IL1A is required for the induction of fibroblast-specific cytokines involved in paracrine signaling between the epidermis and the dermis.28 We discovered that keratinocyte-specific p63 expression is important for the induction of fibroblast-derived cytokines. Further, p63 regulates the keratinocyte-specific KGFR, a trans-membrane receptor that binds fibroblast-derived KGF. Our data are consistent with a previous report linking p63 and KGFR in thymic epithelial cells.34 Most importantly, our data show that p63 is important for regulating target gene expression in the epidermis, and the expression of fibroblast-specific growth factors that promote epidermis development in a paracrine manner. Although our data suggest that p63 regulates paracrine signaling primarily through IL1A production, we cannot rule out the possibility that p63 also regulates the expression of additional secreted factors that function in epithelial–mesenchymal paracrine pathways.
Psoriasis, an epidermal disorder characterized by hyperproliferation, hypodifferentiation, and increased p63 expression,35 further links p63 to paracrine pathways mediated by IL1A. Consistent with our discovery of IL1A being induced by p63, previous studies identified increased expression of KGF and FGF10 in the stroma adjacent to psoriatic lesions.36 On the basis of our data, we would hypothesize that the increased stromal levels of KGF and FGF10 are a direct result of IL1A induction by p63, which is upregulated during psoriatic remodeling.35
We showed that TCF7L1 functions to maintain the undifferentiated state of basal keratinocytes. Ectopically expressed TCF7L1 was sufficient to abrogate calcium-induced NHEK differentiation. In OTC, TCF7L1 depletion was sufficient to prematurely induce differentiation, suggesting that it acts to repress differentiation pathways in human keratinocytes and thus have an important role in tissue homeostasis. These data are consistent with previous studies showing that ectopic TCF3, the mouse TCF7L1 homolog, was sufficient to induce an undifferentiated, proliferative state in mouse keratinocytes.33 There is additional evidence from Xenopus studies that link p63 to TCF7L1. In Xenopus, xTcf3−/− embryos display severely expanded mesodermally derived structures, suggesting that it may act to repress transcriptional programs responsible for mesoderm differentiation.37 We recently identified a role for ΔNp63 as a repressor of mesoderm development during Xenopus embryogenesis.38 Further research is necessary to determine whether ΔNp63 transcriptionally regulates xTcf3 during Xenopus development. Given that previous reports describe p63 as a regulator of proliferation and differentiation in basal epithelial cells,1 our linkage of p63 to TCF7L1 expression in basal epithelial cells provides mechanistic insight into how p63 regulates the maintenance of epithelial progenitor cells.
Lastly, our ChIP dataset proved useful to further identify p63 target genes in a previously published study.19 Overlaying our ChIP dataset with microarray data from their study identified 32 additional p63 target genes not present in this study, such as claudin-1, Jagged-1, and RUNX1, all known p63 target genes, as well as keratinocyte differentiation genes, such as SERPINE1 and PKP1. Ultimately, our ChIP dataset, used in conjunction with microarray analyses, will provide a tiered panel to identify direct versus indirect regulation of target genes by p63 in gene expression analyses.
In conclusion, we show that p63 acts to regulate both cell autonomous and non-autonomous pathways involved in keratinocyte growth and differentiation. Our study provides numerous novel p63 target genes that will be of value to future investigation of transcriptional networks involved in complex epithelia development and differentiated function.
Materials and Methods
Cell culture
Primary NHEKs were obtained and cultured as described previously.3 Primary human dermal fibroblasts were obtained from the Vanderbilt Skin Disease Research core and cultured in Dulbecco's minimal Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. The retrovirus-packaging cell line, Phoenix, was cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. All cells were cultured at 37°C with 5% CO2. Terminal differentiation of NHEKs was performed as described previously.3
Protein lysate preparation and western blot analysis
Protein lysates were prepared from NHEKs as described previously.3 Western analysis was performed as described previously3 with the following primary antibodies: anti-p63 monoclonal antibody 4A4 (Santa Cruz, Santa Cruz, CA, USA), anti-p73 A300 (Bethyl, Montgomery, TX, USA), anti-p53 clone DO-1 (Santa Cruz), anti-TCF7L1 (Santa Cruz), loricrin (Covance, Princeton, NJ, USA), involucrin, and β-actin polyclonal antibody I-19 (Santa Cruz).
Quantitative reverse transcription-PCR
Quantitative reverse transcription-PCR (RT-PCR) and 13 real-time PCR were performed as described previously.4, 13 Primer sequences and PCR conditions are available upon request.
ChIP experiments
Formaldehyde crosslinking and chromatin preparation and ChIP were carried out as described previously.15 For ChIP data generation and semiquantitative ChIP PCR experiments, rapidly growing NHEKs were crosslinked and submitted to GenPathway Inc. (San Diego, CA, USA) according to their FactorPath protocol. The complete p63 ChIP dataset is accessible through Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/).
Microarray analyses
siRNA-mediated disruption of p63 for microarray data generation was performed using the pRetroSuper system as described in ref.4 RNA was harvested and Affymetrix microarray analysis was performed in duplicate as described previously.4 All gene expression data from microarray experiments and genomic location information from the p63 ChIP experiment have been deposited in the National Center for Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo/). The GeneSpring GX software (Agilent, Santa Clara, CA, USA) was used for statistical analyses and transcript annotations. For tumor analysis, publicly available expression data from GSE3524 and GSE2280 were pooled and used to identify tumors with high and low p63 mRNA expression, as measured by mRNA levels. The 82-gene p63 signature was analyzed in these two tumor subgroups to identify genes differentially regulated in tumors relative to p63 expression.
siRNA experiments and immunofluorescence
For validation studies, p63 was depleted using adenoviruses expressing siRNAs targeting the p63DBD or a shuttle control virus as described previously.5 For TA-specific knockdown experiments, TAp63 was depleted using the following siRNA oligonucleotides: siTAp63-1, 5′-CAGAAGATGGTGCGACAAA-3′; siTAP63-2, 5′-GCACACAGACAAATGAATT-3′; and siTAp63-3, 5′-ACTTGAACTTTGTGGATGA-3′. For TCF7L1 knockdown studies, siRNA (Dharmacon Inc., Lafayette, CO, USA) was electroporated into primary NHEKs as per the manufacturer's instructions (Amaxa Biosystems, Walkersville, MD, USA). For immunofluorescence, sections were processed as described previously.39 After primary incubation, sections were incubated with the appropriate secondary antibody (AlexaFluor 488 or 546) (Molecular Probes and Invitrogen, Carlsbad, CA, USA). Nuclei were stained using TOPRO-3 (Invitrogen). Sections were analyzed on a Zeiss Inverted LSM510 confocal microscope (Vanderbilt-Ingram Cancer Center Cell Imaging Shared Resource, Nashville, TN, USA). Primary antibodies used were as follows: loricrin (Covance, Princeton, NJ, USA), involucrin, and filaggrin (Santa Cruz), and E-cadherin (BD Biosciences, Franklin Lake, NJ, USA). For proliferation analyses, the OTCs were incubated with a mixture of BrdU and 5-bromodeoxycytidine (Sigma, St Louis, MO, USA; both 63.5 μM, added to the culture medium) for 18 h before being terminated. DNA incorporation of these nucleotide analogs in replicating cells was detected on cryosections with a monoclonal antibody anti-BrdU antibody (BD Biosciences) after sequential methanol and acetone fixation as described above and 10 min incubation with 1.5 M HCl at room temperature followed by thorough rinsing in PBS. To determine the proliferative indices, BrdU-labeled and all basal cell nuclei were counted in at least three vision fields of three to five sections of each culture specimen.
Organotypic co-culture
OTCs were prepared as described previously.40 Type I collagen was purchased from BD Biosciences. The gels, containing primary fibroblasts and representing the dermal equivalents, were incubated in DMEM supplemented with 10% fetal bovine serum and 50 μg ascorbic acid per ml. Culture medium was changed every 2 days and cultures were harvested for analysis 10 days after lift.
Abbreviations
- ChIP:
-
chromatin immunoprecipitation
- NHEKs:
-
human epidermal keratinocyte
References
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398: 714–718.
Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A . p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999; 398: 708–713.
Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA . The delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay–Wells syndrome-derived mutations. Mol Cell Biol 2003; 23: 2264–2276.
Barbieri CE, Tang LJ, Brown KA, Pietenpol JA . Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res 2006; 66: 7589–7597.
Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 2006; 8: 551–561.
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2: 305–316.
King KE, Ponnamperuma RM, Yamashita T, Tokino T, Lee LA, Young MF et al. DeltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene 2003; 22: 3635–3644.
Helton ES, Zhu J, Chen X . The unique NH2-terminally deleted (deltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the deltaN variant of p63. J Biol Chem 2006; 281: 2533–2542.
Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 1999; 99: 143–153.
McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR et al. Hay–Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet 2001; 10: 221–229.
Li N, Singh S, Cherukuri P, Li H, Yuan Z, Ellisen LW et al. Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells (Dayton, OH) 2008; 26: 1253–1264.
Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR . p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 2004; 18: 126–131.
Barbieri CE, Perez CA, Johnson KN, Ely KA, Billheimer D, Pietenpol JA . IGFBP-3 is a direct target of transcriptional regulation by DeltaNp63alpha in squamous epithelium. Cancer Res 2005; 65: 2314–2320.
Testoni B, Borrelli S, Tenedini E, Alotto D, Castagnoli C, Piccolo S et al. Identification of new p63 targets in human keratinocytes. Cell Cycle 2006; 5: 2805–2811.
Perez CA, Ott J, Mays DJ, Pietenpol JA . p63 consensus DNA-binding site: identification, analysis and application into a p63MH algorithm. Oncogene 2007; 26: 7363–7370.
Ortt K, Raveh E, Gat U, Sinha S . A chromatin immunoprecipitation screen in mouse keratinocytes reveals Runx1 as a direct transcriptional target of DeltaNp63. J Cell Biochem 2008; 104: 1204–1219.
Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR et al. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 2006; 24: 593–602.
Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C et al. New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J 2006; 25: 5105–5116.
Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA . p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 2006; 20: 3185–3197.
Labhart P, Karmakar S, Salicru EM, Egan BS, Alexiadis V, O'Malley BW et al. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc Natl Acad Sci USA 2005; 102: 1339–1344.
Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA . A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol 2008; 28: 5951–5964.
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139: 1327–1341.
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B et al. A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 2009; 137: 87–98.
Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ 2006; 13: 1037–1047.
Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA 2000; 97: 5462–5467.
Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet 2004; 154: 27–35.
O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene 2005; 24: 1244–1251.
Maas-Szabowski N, Fusenig NE . Interleukin-1-induced growth factor expression in postmitotic and resting fibroblasts. J Invest Dermatol 1996; 107: 849–855.
Maas-Szabowski N, Shimotoyodome A, Fusenig NE . Keratinocyte growth regulation in fibroblast cocultures via a double paracrine mechanism. J Cell Sci 1999; 112 (Part 12): 1843–1853.
Aaronson SA, Rubin JS, Finch PW, Wong J, Marchese C, Falco J et al. Growth factor-regulated pathways in epithelial cell proliferation. Am Rev Resp Dis 1990; 142 (Part 2): S7–10.
Marchese C, Rubin J, Ron D, Faggioni A, Torrisi MR, Messina A et al. Human keratinocyte growth factor activity on proliferation and differentiation of human keratinocytes: differentiation response distinguishes KGF from EGF family. J Cell Physiol 1990; 144: 326–332.
Merrill BJ, Gat U, DasGupta R, Fuchs E . Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev 2001; 15: 1688–1705.
Nguyen H, Rendl M, Fuchs E . Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 2006; 127: 171–183.
Candi E, Rufini A, Terrinoni A, Giamboi-Miraglia A, Lena AM, Mantovani R et al. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci USA 2007; 104: 11999–12004.
Shen CS, Tsuda T, Fushiki S, Mizutani H, Yamanishi K . The expression of p63 during epidermal remodeling in psoriasis. J Dermatol 2005; 32: 236–242.
Kovacs D, Falchi M, Cardinali G, Raffa S, Carducci M, Cota C et al. Immunohistochemical analysis of keratinocyte growth factor and fibroblast growth factor 10 expression in psoriasis. Exp Dermatol 2005; 14: 130–137.
Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV et al. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development (Cambridge, UK) 2004; 131: 263–274.
Barton CE, Tahinci E, Barbieri CE, Johnson KN, Hanson AJ, Jernigan KK et al. DeltaNp63 antagonizes p53 to regulate mesoderm induction in Xenopus laevis. Dev Biol 2009; 329: 130–139.
Muffler S, Stark HJ, Amoros M, Falkowska-Hansen B, Boehnke K, Buhring HJ et al. A stable niche supports long-term maintenance of human epidermal stem cells in organotypic cultures. Stem Cells (Dayton, OH) 2008; 26: 2506–2515.
Cerezo A, Stark HJ, Moshir S, Boukamp P . Constitutive overexpression of human telomerase reverse transcriptase but not c-myc blocks terminal differentiation in human HaCaT skin keratinocytes. J Invest Dermatol 2003; 121: 110–119.
Acknowledgements
This work was supported by NIH Grants CA70856 and CA105436 (JAP), ES00267 and CA68485 (core services), and NIH Training Grant T32CA078136-08 (CEB). We thank Dr Christopher Barbieri for generating the shp63DBD retrovirus and Dr Leif Ellisen for providing the siRNA adenoviruses. The Krt2 antibody was kindly provided by Dr Lutz Langbein (DKFZ). We would like to thank members of the Pietenpol laboratory for critical reading of the manuscript and helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Disease website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Barton, C., Johnson, K., Mays, D. et al. Novel p63 target genes involved in paracrine signaling and keratinocyte differentiation. Cell Death Dis 1, e74 (2010). https://doi.org/10.1038/cddis.2010.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2010.49
Keywords
This article is cited by
-
Structural diversity of p63 and p73 isoforms
Cell Death & Differentiation (2022)
-
p63 suppresses the ability of pregnancy-identified mammary epithelial cells (PIMECs) to drive HER2-positive breast cancer
Cell Death & Disease (2021)
-
Human papillomavirus E7 induces p63 expression to modulate DNA damage response
Cell Death & Disease (2018)
-
Retinoic acid and BMP4 cooperate with p63 to alter chromatin dynamics during surface epithelial commitment
Nature Genetics (2018)
-
Enlightening discriminative network functional modules behind Principal Component Analysis separation in differential-omic science studies
Scientific Reports (2017)