Abstract
Background:
Lung cancer is a leading cause of cancer-related mortality worldwide and non-small-cell lung carcinoma (NSCLC) is responsible for almost 80% of lung cancer-related deaths. Identifying novel molecules that can repress the invasiveness and metastasis of lung cancer will facilitate the development of new antilung cancer strategies. The aim of this study is to determine the roles of NUAK1 (a downstream of Akt) and miR-204 in the invasiveness and metastasis of NSCLC and to reveal the correlation between NUAK1 and miR-204.
Methods:
The expression of NUAK1 in primary human NSCLC tissues was evaluated by immunohistochemistry. Real-time PCR was employed to measure the expression level of miR-204. The effect of NUAK1 and miR204 on the prognosis of NSCLC patients was evaluated by log-rank test. The siRNA transfection was used to manipulate the expression levels of NUAK1 and miR204 in cancer cells. Chemotaxis assay, Scratch assay, and Matrigel invasion assay were performed to evaluate the migration and invasion of cells. Cellular F-actin measurement was used to measure F-actin polymerisation in lung cancer cells. Western blot was used to detect the expression levels of corresponding proteins. The Luciferase assay and RNA immunoprecipitation were used to confirm the actual binding site of miR-204 to 3′UTR of NUAK1.
Results:
Increased expression of NUAK1 is correlated with the invasiveness and metastasis of human NSCLC. Knockdown of NUAK1 inhibited cell migration and invasion. In addition, this study showed that NUAK1 influenced mTOR phosphorylation and induced the phosphorylation of p70S6K1 and eukaryotic initiation factor 4E-binding protein1 (4E-BP1), two downstream targets of mTOR in NSCLC cells. At the same time, decreased expression of miR-204 promoted NSCLC progression and, contrarily, manipulated upregulation of miR-204-inhibited cell migration and invasion. There is clinical relevance between miR-204 downregulation and NUAK1 upregulation in human NSCLC. Furthermore, we found that miR-204 inhibited NSCLC tumour invasion by directly targeting and downregulating NUAK1 expression. Finally, our data suggested that the downregulation of miR-204 was due to hypermethylation of its promoter region.
Conclusions:
Our results indicate that NUAK1 is excessively expressed in NSCLC and plays important roles in NSCLC invasion. The miR-204 acts as a tumour suppressor by inhibiting NUAK1 expression in NSCLC. Both NUAK1 and miR-204 may serve as potential targets of NSCLC therapy.
Similar content being viewed by others
Main
Lung cancer is a leading cause of cancer-related mortality worldwide and non-small-cell lung carcinoma (NSCLC) is responsible for almost 80% of lung cancer-related deaths (Buyukcelik et al, 2004; Jemal et al, 2011). Despite recent improvements in chemotherapy and molecular-targeted therapy, the outcome of lung cancer remains poor. The invasiveness and metastasis of tumour cells are critical challenges in the clinical management of NSCLC. Identifying novel molecules that can repress the invasiveness and metastasis of lung cancer cells will facilitate the development of new antilung cancer strategies.
The NUAK1 is a novel member of the AMP-activated protein kinase (AMPK) catalytic subunit family and is directly activated by Akt (Suzuki et al, 2004). Recent studies provide evidences that NUAK1 is closely involved in tumour progression. NUAK1 has been found to induce tumour cell survival during nutrient starvation in an Akt-dependent manner (Suzuki et al, 2003). High expression of NUAK1 was involved in tumour progression of colon cancer clinically (Kusakai et al, 2004). In addition, NUAK1 enhanced the invasive and metastatic potential of breast cancer cells under regulation by Akt (Chang et al, 2012). Furthermore, in a recent study in our laboratory, we demonstrated that upregulation of NUAK1 promoted glioma invasion (Lu et al, 2013). Although these studies indicate the importance of NUAK1 in cancer progression, its roles in NSCLC still remain to be investigated.
MicroRNAs (miRNAs) are small, noncoding single-stranded RNAs that originate from encoding genes in nucleus. MicroRNAs downregulate gene expression by binding partially and complementarily to the 3′-untranslated regions (UTRs) of target mRNAs in post-transcriptional level, leading to mRNA degradation or inhibition of translation (Lagos-Quintana et al, 2001; Lim et al, 2005). Approximately 20–30% of all genes are targeted by miRNAs and a single miRNA may target as many as 200 genes (Krek et al, 2005). Although their biological functions remain largely unknown, recent studies suggest that miRNAs are involved in cancer progression and metastasis by targeting oncogenes and tumour suppressor genes (Garzon et al, 2009; Ryan et al, 2010). Many miRNAs are known to be up- and/or downregulated in a variety of cancers and can have either tumour-promoting or tumour-suppressing effects (Volinia et al, 2006; Ventura and Jacks, 2009; Ventura et al, 2009). In addition, a growing body of evidences indicate that certain miRNAs can function as potential biomarkers for cancer diagnosis, progress, and response to treatment (Lu et al, 2005; Garzon and Marcucci, 2012).
The human miR-204 is located in the 7q32 region and is transcribed from the cluster of miR-183 family. There is emerging knowledge about the roles of miR-204 in cancers (Chen et al, 2013). In fact, recent advances in miRNA-related researches have shown that miR-204 has important roles in tumourigenesis, including the regulation of carcinogenesis in gastric cancer (Lam et al, 2011; Sacconi et al, 2012; Zhang et al, 2013), squamous cell carcinoma of the head and neck (Lee et al, 2010), peripheral nerve sheath tumours (Gong et al, 2012), bladder cancer (Pignot et al, 2013), and the migration and invasion of endometrial cancer cell lines (Chung et al, 2012). Imam et al (2012) reported that chromosomal loci containing miR-204 is frequently lost, resulting in its lower expression in multiple cancers. The miR-204 acted as a potent suppressor of tumour growth and metastasis for ovarian cancers, breast cancers, and pediatric renal tumours (Imam et al, 2012). On the other hand, however, miR-204 showed tumour-promoter activity in human prostate cancer (Turner et al, 2011). The functions and experimental targets of miR-204 varied depending on cellular types used in experiments, suggesting a significant influence of the molecular background on miRNA target selection. Till now, the roles of miR-204 in NSCLC have never been described and this prompted us to identify and validate the role of miR-204 in NSCLC.
In the current study, for the first time, we demonstrated that NUAK1 played a key role in NSCLC progression. In addition, we found a downregulation of miR-204 in NSCLC tissues and cells. Functional assays showed that the expression of miR-204 was inversely correlated with the expression of NUAK1 and the invasive potential of NSCLC. Moreover, NUAK1 was confirmed to be a direct target gene of miR-204.
Materials and Methods
Patients and tissue specimens
This study was conducted on a total of 207 paraffin-embedded NSCLC samples that were histopathologically diagnosed at the Department of Pathology of Weifang Medical University from 2002 to 2006. The histological characterisation and clinicopathological staging of the samples were determined according to the Union for International Cancer Control (UICC) Tumour–Node–Metastasis (TNM) classification. For western blot, 20 pairs of selected frozen (liquid nitrogen) NSCLC tissues and adjacent nontumour tissues (ANTs) were evaluated. For the use of these clinical materials for research purpose, prior patients’ consents and approval from the Institutional Research Ethics Committee were obtained. Clinical information of the specimens is described in detail in Supplementary Table 1.
Immunohistochemistry
Primary antibody used for immunostaining was the NUAK1 polyclonal antibody (sc-130117, 1 : 100 dilution; Santa Cruz, Dallas, TX, USA). Briefly, slides were deparaffinised in xylene and transferred through two changes of 100% ethanol. For antigen retrieval, the slides were boiled in a pressure cooker at maximum heat for 2 min containing 0.01 mol l−1 sodium citrate (pH 6.0) and cooled for 30 min at room temperature. Endogenous peroxidase activity was blocked in 0.3% H2O2 for 10 min at 37 °C. After incubation, the slides were washed 3 times in PBS for 3 min each. Then slides were incubated with the primary antibody overnight at 4 °C. Routine controls using PBS instead of the primary antibody were included to verify specificity. After washing with PBS, the bound primary antibody was detected by Biotinylated Goat Anti-Rabbit IgG (H+L) (1 : 200 dilution; Vector Laboratories, Burlingame, CA, USA) and the chromogenic substrate 3,3-diaminobenzidine tetrahydrochloride (DAB). The specimens were counterstained with haematoxylin, mounted, and examined by light microscopy (Olympus IX71, Tokyo, Japan).
Immunohistochemical evaluation
The degree of immunostaining of the sections was viewed and scored separately by two independent investigators who were blinded to the histopathologic features and patient data of the samples. The results were evaluated by an immunohistochemical score (IHS) (Li et al, 2008). The IHS was calculated by combining an estimate of the percentage of immunoreactive cells (quantity score) with an estimate of the staining intensity (staining intensity score) as follows: no staining is scored as 0, 1–10% of cells stained scored as 1, 11–50% as 2, 51–80% as 3, and 81–100% as 4. The percentage of positive cells was calculated by counting more than 1000 cancer cells in randomly selected high-power fields ( × 400). Staining intensity was rated on a scale of 0–3, with 0=negative, 1=weak, 2=moderate, and 3=strong. When there was multifocal immunoreactivity and there were significant differences in staining intensities between foci, the average of the least intense and most intense staining was recorded. The raw data were converted to IHS by multiplying the quantity and staining intensity scores. Theoretically, the scores could range from 0 to 12. The HIS score of 9–12 was considered strong immunoreactivity, 5–8 was considered moderate, 1–4 was considered weak, and 0 was scored as negative. In addition, 0–4 was regarded as low expression and 5–12 was regarded as high expression. The NSCLC patients were divided into two groups based on NUAK1 expression level, namely, the low NUAK1 expression group and the high NUAK1 expression group for clinical survival analysis.
Cell culture
Lung cancer cell lines, including SPC, A549, NCI-H1299, and Calu1, were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in RPMI-1640 (SH30809.01B, HyClone, Logan, UT, USA) and supplemented with 10% fetal bovine serum (SH30070.03, HyClone) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 °C in a humidified atmosphere of 95% air and 5% CO2. The BEAS-2B immortalised human bronchial epithelial cell line (Shanghai Institutes of Biological Sciences, Shanghai, China) was cultured in LHC-9 medium as instructed by the provider. The chemotaxis chambers and membranes were from Neuroprobe (Gaithersburg, MD, USA). The human EGF (epidermal growth factor) was purchased from R&D Systems (Minneapolis, MN, USA). To inhibit hypermethylation in the promoter region of miR-204 gene, DNA methyltransferase inhibitor 5-Aza-2′-deoxyazacytidine (5-aza-dC; Sigma, St Louis, MO, USA) was used to treat the forementioned cell lines (Ambros, 2008). Cells were seeded in 10 cm dishes (1 × 106 cells per dish) 24 h before drug treatment. The cells were treated with 5 μ M 5-aza-dC for 72 h and dimethylsulphoxide (DMSO; Sigma) was used as negative control.
Plasmid construction and transfection
Cells were plated in a 35 mm dish for 24 h before transfection into the complete medium. The transfection was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For transient transfections, two Stealth siRNAs against human NUAK1 (#1: 5′-GAAGTTATGCTTTATTCAC-3′, #2: 5′-CATCCTCTGATTCTAGGTG-3′) and a scrambled siRNA were synthesised by Invitrogen. BLOCK-iT Fluorescent Oligo was used to examine the transfection efficiency (Invitrogen). The transient transfected cells were named SiNUAK1#1/H1299, SiNUAK1#2/H1299, and Scr/H1299 cells. At 72 h after transfections, cells were harvested and used for further experiments. For stable transfection, a siRNA expression plasmid containing target sequence (5′-CATCCTCTGATTCTAGGTG-3′) and a vector containing a scrambled sequence were selected by using 600 μg ml−1 neomycin. The stable transfected cells are named siNUAK/H1299 and Scr/H1299 cells for subsequent studies.
The 3′UTRs of NUAK1 containing predicted miR-204 target sites were amplified by PCR from A549 cell genomic DNA and cloned into pcDNA3.1 expression vector (GenePharma Company, Shanghai, China). The A549 cells were transfected with pcDNA3.1-NUAK1 plasmid or pcDNA3.1 vector using Lipofectamine 2000 (Invitrogen) following the protocol. After 24 h of transfection, cells were trypsinised, diluted, and reseeded into 10 cm culture dishes. Stable transfected cells were obtained by using selection medium (culture medium with 700 μg ml−1 G418). Single cell clones were isolated for clone expansion. Stable transfected cell clones were named A549/NUAK1 and A549/CON cells. The cells were maintained and passaged in culture medium with G418 (400 μg ml−1). The 3′UTRs of NUAK1 were amplified and then cloned into the downstream of the luciferase gene in a modified pGL3 control vector (Promega, Madison, WI, USA) (Li et al, 2013).
The miR-204 mimics, miR-204 inhibitor oligonucleotides, miR-204 mutants, and corresponding control oligonucleotide mimics (NC) were synthesised by GenePharma company (Shanghai, China). A DNA fragment containing the hsa-miR-204 precursor with 300 bp flanking sequence of each side was amplified into retroviral transfer plasmid pMSCVpuro (GenePharma). Retroviral production and infection were performed as described previously. Following transduction, puromycin (1.5 μg ml−1) was used as a selection antibiotic to select the infected cells for 10 days (Li et al, 2013). Stable transfected cell clones were named miR-204/H1299, miR-204 inhibitor/H1299, miR-204 mutant/H1299, and miR-NC/H1299 or miR-204/A549, miR-204 inhibitor/A549, miR-204 mutant/A549, and miR-NC/A549 cells, respectively.
RNA extraction and real-time quantitative PCR
Total miRNAs of cultured cells, 15 surgically resected fresh NSCLC tissues and 80 of 207 randomly chosen paraffin-embedded, archived clinical NSCLC specimens were extracted using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) and RecoverAll Total Nucleic Acid Isolation kit (Ambion) according to the manufacturer’s instruction. Complementary DNA was synthesised with 5 ng of total RNA using the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), and the expression levels of miR-204 were quantified with the miRNA-specific TaqMan MiRNA Assay Kit (Applied Biosystems) (Cai et al, 2013). The corresponding 95 NSCLC patients were divided into two groups based on miR-204 expression level, namely, the low-miR-204 expression group (below the median value) and the high-miR-204 expression group (above the median value), for clinical survival analysis.
Bisulphite genomic sequencing
Genomic DNAs from BEAS-2B immortalised human bronchial epithelial cell line, NSCLC cell lines, and clinical specimens were bisulphite modified with the Epitect Bisulphite Kit (Qiagen, Duesseldorf, Germany). Bisulphite-treated DNAs were amplified with BSP (bisulphite-sequencing PCR) primers located in the miR-204 promoter (Ying et al, 2013). The PCR products were cloned using the pGEM-T Easy Vector System (Promega). Plasmids from single colonies were purified and sequenced.
Western blot assay
For western blot, cells or tissues were directly lysed in 1 × SDS sample buffer. Protein samples were separated by SDS–PAGE, transferred to polyvinylidene difluoride membranes (Millipore Inc., Billerica, MA, USA), immunoblotted using the appropriate primary and the HRP-conjugated secondary antibodies, and were visualised by using enhanced chemiluminescence reagents ECL (Pierce, Rockford, IL, USA). Blotted membranes were stripped and reblotted with an anti-β-actin monoclonal antibody (Cell Signaling, Danvers, MA, USA) as loading control. The intensities of bands in western blots were quantified by densitometry analysis using AlphaImager HP (Alpha Innotech, San Leandro, CA, USA) and NIH Image J software (Rockville, MD, USA). Western blot data shown in the paper are representatives from three independent experiments. The following commercial antibodies were used in this study: NUAK1 (4458, 1 : 1000 dilution; Cell Signaling), p-cofilin (sc-21867-R, 1 : 1000 dilution; Santa Cruz), p-LIMK (3841, 1 : 1000 dilution; Cell Signaling), LIMK (3842, 1 : 1000 dilution; Cell Signaling), MMP-2 (13132, 1 : 1000 dilution; Cell Signaling), MMP-9 (3852, 1 : 1000 dilution; Cell Signaling), MT-MMP (3852, 1 : 1000 dilution; Cell Signaling), p-mTOR (5536, 1 : 1000 dilution; Cell Signaling), p-4E-BP1 (2855, 1 : 1000 dilution; Cell Signaling), p-P70S6K (9234, 1 : 1000 dilution; Cell Signaling), β-actin (4970, 1 : 1000 dilution; Cell Signaling), and HRP-linked Anti-rabbit IgG antibody (7074, 1 : 2000 dilution; Cell Signaling).
Chemotaxis assay
Chemotaxis assays were performed by using transwell chambers as described previously (Zhang et al, 2009a). Briefly, the chemoattractant (EGF) was loaded into the lower chemotaxis chamber and 5 × 105 cells per ml cells suspended in the binding medium (RPMI-1640, 0.1% BSA, and 25 mM HEPES) were added to the upper chambers. The polycarbonate filter (8 μm pore size) was pretreated with 10 μg ml−1 fibronectin overnight, dried in air, and inserted between the upper and lower chambers. Then, the chamber was incubated at 37 °C in 5% CO2 for 3 h. The filter membrane was then rinsed, fixed, and stained. The numbers of migrating cells were counted at × 400 in three separate fields by light microscope.
Scratch assay
The Scr/H1299 and SiNUAK1/H1299 cells were plated in 35 mm dishes for 2 days to grow into a monolayer. Then, it was lined out with an even trace in the middle using a 10-μl pipette tip. The cells were then incubated at 37 °C in 5% CO2 within an appropriate time and the distance of the wounds was measured under a light microscope. All samples were tested in triplicate, and the data were expressed as mean±s.d.
Cellular F-actin measurement
The F-actin content was detected as described previously (Zhang et al, 2009b; Li et al, 2013). In brief, the cells were suspended at 4 × 105 per ml in medium supplemented with 2% (w/v) BSA and 10 mM HEPES (pH 7.2) and preincubated for 3 h at 37 °C. The cells were followed by the stimulation of 10 ng ml−1 EGF at 37 °C at different time points. The cells were then fixed with 4% paraformaldehyde for 10 min, permeabilised with 0.1% Triton X-100 for 30 min, and incubated with Alexa-fluro 568 phalloidin in F-actin buffer (10 mmol l−1 HEPES, 20 mmol l−1 KH2PO4, 5 mmol l−1 EGTA, 2 mmol l−1 MgCl2, PBS, pH 7.4) at room temperature for 60 min. The cells were washed 5 times. F-actin content was measured with a microplate fluorescence reader with an excitation wavelength of 578 nm and an emission wavelength of 600 nm. The relative F-actin content over different time periods was calculated by the following equations: F-actin Δt/F-actin 0=fluorescence Δt/ fluorescence 0.
Matrigel invasion assay
A Boyden chamber invasion assay was performed as previously described (Albini et al, 1987; Lu et al, 2013). The cells were suspended in serum-free medium at a final concentration of 4 × 105 per ml and incubated at 37 °C for 30 min. Then, the cells, in the presence or absence of 10 ng ml−1 EGF, were promptly added to a 35 mm dish containing dried fibronectin-coated glass coverslips. After an incubation of 24 h, the cells were washed gently twice with cold PBS and fixed. The attached cells were counted under a light microscope at × 200.
Gelatin zymography
Cells transfected with control or NUAK1 siRNA were treated with 10 ng ml−1 EGF for 12 h in serum-free medium. The conditioned medium was collected and analysed on 10% SDS–polyacrylamide gel incorporated with 0.1% gelatin (Das et al, 2011). The gel was stained with 0.2% Coomasie Brilliant Blue in 40% isopropanol for 25 min and destained in 7% glacial acetic acid. In our experiments, MMP-9 and MMP-2 were the only gelatinases detected at 92 and 72 kDa, respectively, and were visualised as decolourised bands over blue background.
Luciferase reporter assay
Cells (3.5 × 104) were seeded in triplicates in 48-well plates and allowed to settle for 24 h. Then, 100 ng of pGL3-NUAK1-3′UTR, plus 1 ng of pRL-TK Renilla plasmid (Promega), were transfected using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s recommendation. Luciferase and Renilla signals were measured at 48 h after transfection using the Dual Luciferase Reporter Assay Kit (Promega) according to a protocol provided by the manufacturer.
RNA immunoprecipitation (RIP)
Co-immunoprecipitation (Co-IP) of miRNP with anti-Ago1 (Abcam, Cambridge, MA, USA) or IgG (Sigma) was performed as previously described (Tan et al, 2009). RNA co-immunoprecipitated with anti-Ago1 or IgG antibodies was extracted using TRIzol LS (Invitrogen) as described in a previous publication (Cheng et al, 2008). Total RNA was subjected to DNA digestion as described above.
Xenografted tumour model in vivo and immunohistochemistry (IHC)
To investigate whether NUAK1 and miRNA-204 could affect NSCLC metastasis in vivo, 5 × 105 corresponding cells (Scr/H1299, SiNUAK1/H1299, miR-NC/H1299, or miR-204/H1299) were implanted into 4-week-old male nude mice by tail vein injection individually with 10 mice per group. To investigate the molecular mechanism that mediates the downregulation of miR-204 in NSCLC tumours, H1299 cells (5 × 105) were implanted individually into 20 4-week-old male nude mice by tail vein injection. The mice were randomly divided into two groups with 10 mice per group. In the first week after tumour inoculation, 200 μl 5-aza-dC (400 μ M) or DMSO (negative control) was intravenously injected through tail vein into corresponding nude mice group once a day for 1 week (Yoo and Jones, 2006). All animals received humane care according to the criteria outlined in the Guide (Publication No. 85-23, revised 1985) for the care and use of laboratory animals. The study protocol was approved by the Review Committee for the Use of Human or Animal Subjects of Weifang Medical University. The animals were killed and autopsied 4 weeks after tumour inoculation. Metastasis formation was determined by macroscopic observation and histological examination of lung tissues. The lung tissues were serially cut into 5 μm slices, and every tenth section was stained with H&E to evaluate the presence or absence of lung metastasis. Two independent pathologists calculated the number of metastases in whole lungs. Furthermore, metastatic lung tissues were subjected to western blot for NUAK1 expression.
Statistical analysis
All statistical analyses were carried out using the SPSS 13.0 statistical software package (Chicago, IL, USA). Comparisons between groups for statistical significance were performed with a two-tailed paired Student’s t-test. The χ2 test was used to analyse the relationship between NUAK1 or mir204 expression and clinicopathologic characteristics. Survival curve was plotted using the Kaplan–Meier method and compared by the log-rank test. P<0.05 was considered statistically significant in all cases.
Results
Increased expression of NUAK1 in primary human NSCLC and NSCLC cell lines
To investigate whether NUAK1 protein is overexpressed in clinical samples of malignant lung tumour tissues, we performed immunohistochemical analysis. We mainly assessed the corelation between NUAK1 expression and clinicopathologic features of patients with lung squamous cell carcinomas and adenocarcinomas. As for lung tumour tissues, except for few samples that showed nucleolus staining, most lung tumour tissues showed strong cytoplasmic staining with NUAK1. On the contrary, in the normal lung tissues, few cells showed positive staining with NUAK1 (Figure 1A). Among all paraffin-embedded lung cancer tissue samples, positive NUAK1 expression rates are as following: 74.76% (77 out of 103) for lung squamous cell carcinomas and 71.15% (74 out of 104) for lung adenocarcinomas. Inversely, among 40 paraffin-embedded normal lung tissues, positive NUAK1 expression rate is only 22.5% (9 out of 40), markedly lower than that of lung cancer tissue samples. In addition, there is no statistical difference of NUAK1 expression between lung squamous cell carcinomas and adenocarcinomas (Supplementary Table 1).
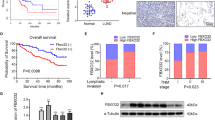
The expression of NUAK1 in NSCLC tissues and NSCLC cell lines. (Aa) Squamous cell carcinoma without lymph node metastasis, (Ab) squamous cell carcinoma with lymph node metastasis, (Ac) adenocarcinoma without lymph node metastasis, and (Ad) adenocarcinoma with lymph node metastasis were examined by immunohistochemical staining. Scale bar: 10 μm. (B, left) Expression of NUAK1 protein in cultured human bronchial epithelial cell line (BEAS-2B) and NSCLC cell lines (SPC-A1, A549, H1299, and Calu1). (B, right) Expression of NUAK1 protein in paired NSCLC tissues (T) and adjacent nontumour tissues (ANTs), with each pair obtained from the same patient. β-Actin was used as a loading control. The results were from a representative of at least three repeated experiments. (C) Kaplan–Meier analysis of correlation between NUAK1 level and survival of NSCLC patients in stage I–II subgroup. (D) Kaplan–Meier analysis of correlation between NUAK1 level and survival of NSCLC patients in stage III–IV subgroup. (E) Kaplan–Meier analysis of correlation between NUAK1 level and overall survival of NSCLC patients with low (n=87) and high (n=120) NUAK1 expression. P-values were calculated by the log-rank test, P<0.05 was considered statistically significant.
To further reveal the role of NUAK1 in human NSCLC cancer, western blot was performed to measure NUAK1 expression in a series of NSCLC cell lines and the immortalised human bronchial epithelial cell line BEAS-2B. The results showed that all the NSCLC cell lines have higher expression of NUAK1 than BEAS-2B cell line (Figure 1B). In addition, western blot analyses on 20 pairs of selected human frozen (liquid nitrogen) NSCLC tissues and adjacent ANTs showed that the expression of NUAK1 was significantly increased in cancer tissues compared with matched adjacent noncancerous tissues (Figure 1B). These results clearly showed notable upregulation of NUAK1 in both clinical primary NSCLC cancer tissues and human NSCLC cell lines.
Increased expression of NUAK1 is correlated with metastasis and progression of human NSCLC
To further understand the influences of NUAK1 on NSCLC, we analysed the relationship between NUAK1 expression and clinicopathologic features of the patients. We found that NUAK1 expression was strongly correlated with lymph node metastasis (P=0.009) and distant metastasis (P=0.001) in both aforementioned types of lung cancers respectively, but not with tumour size, age, gender, and histological grade (Supplementary Table 1). Further quantitative analysis indicated that the average mean score of NUAK1 staining was obviously increased in lung cancer tissues with lymph node and distant metastasis (Figure 1A), confirming that high expression of NUAK1 was correlated with metastasis of human NSCLC.
Moreover, Kaplan–Meier analysis using log-rank test was performed to assess the potency of NUAK1 expression on survival of patients. We examined the prognostic value of NUAK1 expression in different subgroups of patients stratified in relation to the UICC staging. Significant correlation between high NUAK1 expression and shorter overall survival time was found in UICC staging subgroups. Patients with tumours exhibiting high NUAK1 expression had significantly shorter overall survival than those with low expression of NUAK1 in either staging I+II subgroup (n=101; P=0.002, log-rank; Figure 1C) or staging III+IV subgroup (n=106; P=0.006, log-rank; Figure 1C). The median survival time of staging I+II subgroup with low NUAK1 expression (43±3.44 months, 95% confidence interval: 36.256–49.744) was significantly longer than that of patients with high NUAK1 expression (29±2.560 months, 95% confidence interval: 23.981–34.019). Similarly, the median survival time of staging III+IV subgroup with low NUAK1 expression (27±2.596 months, 95% confidence interval: 21.911–32.089) was significantly longer than that of patients with high NUAK1 expression (18±1.985 months, 95% confidence interval: 14.109–21.891). The results showed that NUAK1 could be a valuable prognostic marker for lung cancer patients at all disease stages.
Reduction of NUAK1 impaired NSCLC cell migration
In the attempt to understand the biological function of NUAK1, we performed cell chemotaxis assay and scratch assay. As shown in Figure 1B, A549 cells showed relatively lower NUAK1 protein expression, whereas H1299 cells showed relatively higher NUAK1 expression. We selected these two cell lines to verify our hypothesis. PcDNA3.1-NUAK1 plasmid was stably transduced into A549 cell lines to generate A549/NUAK1 cells (Figure 2A). Ectopic NUAK1 expression of A549/NUAK1 cells showed increased chemotaxis compared with control A549/CON cells, whereas there was no obvious difference between parental A549 and A549/CON cells (Figure 2C). In addition, we got the RNA interference (RNAi)-mediated NUAK1 knockdown H1299 cells (SiNUAK1/H1299) and the RNAi control H1299 cells (Scr/H1299) (Figure 2D). The robust chemotaxis of SiNUAK1/H1299 cells decreased compared with Scr/H1299 cells when induced by EGF (Figure 2E). These results indicated that NUAK1 played an important role in the chemotaxis of lung cancer cells. The change in cell proliferation, however, did not interfere with the chemotaxis of cells because it only took <3 h to complete the chemotaxis assay in this study, shorter than the doubling time (Figure 2B). In addition, a scratch assay has also been used as an assay for directional movement. When a scratch was created in the fluent monolayer cells, there was a significant difference between the migration distance of Scr/H1299 and SiNUAK1/H1299 cells, and it took a longer time for SiNUAK1/H1299 cells to fill the gap, further supporting that NUAK1 played an important role in directional migration (Figure 2F).
In NSCLC cells, NUAK1 promoted migration and actin polymerisation. (A) Expression of NUAK1 protein in A549, A549/CON, and A549/NUAK1 cells was detected by western blot; β-actin was used as a loading control. The results were representatives from at least three repeated experiments. (B) Cell proliferation of A549/CON and A549/NUAK1 cells. Each data point was an average of triplicate assays. (C) Comparison of chemotactic responses with EGF stimulation in A549, A549/CON, and A549/NUAK1 cells. Columns indicate means of triplicate measurements, and bars indicate s.d. *P<0.05 (two-way ANOVA). (D) Expression of NUAK1 in Scr/H1299, SiNUAK1#1/H1299, and SiNUAK1#2/H1299 cells was detected by western blot. β-Actin was used as a loading control. (E) Comparison of chemotastic responses of H1299, Scr/H1299, and SiNUAK1/H1299 cells to EGF stimulation. Columns indicate means of triplicate measurements and bars indicate s.d. *P<0.05 (two-way ANOVA). (F) Comparison of cell directional movement of Scr/H1299 and SiNUAK1/H1299 cells by scratch assay. EGF, 10 ng ml−1. Each data point was an average of triplicate assays.
Knockdown of NUAK1 impaired the EGF-induced F-actin polymerisation in H1299 cells
The ligand-induced transient F-actin assembly plays a vital role in cellular chemotactic capacity (Zhang et al, 2010). We performed F-actin polymerisation assay to examine whether the reduction of NUAK1 impaired F-actin polymerisation of H1299 cells. The EGF elicited a transient actin polymerisation at 15 and 60 s in Scr/H1299 cells. In SiNUAK1/H1299 cells, the actin polymerisation in response to EGF stimulation was significantly inhibited, indicating that NUAK1 played a key role in cytoskeleton rearrangement (Supplementary Figure 1A).
As a serine kinase, LIM kinase 1 (LIMK1) specifically phosphorylates members of the cofilin/actin depolymerising factor family and is implicated in the regulation of actin cytoskeletal dynamics. F-actin dynamics is regulated by phosphorylating cofilin at Ser3 that is critical for cell migration and chemotaxis (Arber et al, 1998; Ghosh et al, 2004; Nishita et al, 2005). To test the hypothesis that NUAK1 mediates EGF-induced activation of LIMK1 and cofilin, which in turn regulates actin polymerisation, we examined the evidence for LIMK1 and cofilin activation. As shown in Supplementary Figure 1B, phosphorylation of LIMK1 and cofilin was suppressed in NUAK1 knockdown cells. This result indicates that NUAK1 plays an important role in EGF-induced cofilin recycling and actin polymerisation.
Reduction of NUAK1 impaired NSCLC cell invasion
To further explore the effect of NUAK1 on invasion, we employed a Transwell invasion assay. The EGF (10 ng ml−1) was used as a chemoattractant to stimulate H1299 cells to penetrate through the matrigel and migrate through the filters. Compared with control cells, SiNUAK1/H1299 cells showed dramatically reduced invasive ability Supplementary Figure 2A). We detected the rate of cell proliferation in both SiNUAK1/H1299 and Scr/H1299 cells, and the results showed that knockdown of NUAK1 did not significantly influence cell proliferation.
Knockdown of NUAK1 is relevant to decreased activation of MMP-2 and MMP-9
We have proved that knockdown of NUAK1 can reduce the invasive ability of NSCLC cells. As MMPs are known to be involved in NSCLC invasion (Takemoto et al, 2007; Weng et al, 2013; Xu et al, 2013), we detected the expression of MMPs in SiNUAK1/H1299 and control cells. As shown in Supplementary Figure 2B, western blotting assay showed that there was no obvious difference in the expressions of MMP-2 and MMP-9 between SiNUAK1/H1299 and Scr/H1299 cells. On the other hand, MT1-MMP, which facilitates the activation of MMP-2 and MMP-9, had a remarkable lower expression in SiNUAK1/H1299 cells. Furthermore, the active forms of MMP-2 and MMP-9 in cell culture medium were detected by gelatin zymography. Interestingly, the amounts of active MMP-2 and MMP-9 were downregulated in SiNUAK1/H1299 cells (Supplementary Figure 2C). These results are consistent with our previous research on glioma cell lines and strongly suggest that NUAK1 stimulates MMP-2 and MMP-9 activation via MT1-MMP production.
The mTOR pathway is pivotal in cancer progression, and activation of mTOR is associated with cancer cell survival, proliferation, migration, and invasion (McAuliffe et al, 2010; Imam et al, 2012). Previous studies showed that NUAK1 promoted migratory and invasive properties of glioma through Akt-mTOR activation (Lu et al, 2013). In order to examine whether NUAK1 influences mTOR phosphorylation and induces the phosphorylation of p70S6K1 and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1), two downstream targets of mTOR in NSCLC cell (Khan et al, 2012), western blot was performed to analyse the EGF-induced phosphorylation of mTOR pathway. As shown in Supplementary Figure 2D, EGF-induced phosphorylation of mTOR, 4E-BP1, and P70S6K was significantly decreased in SiNUAK1/H1299 cells compared with Scr/H1299 cells.
MiR-204 directly targets NUAK1
Emerging data imply that miRNAs can regulate tumourigenesis and malignant progression by functioning as tumour suppressors or oncogenes. In mammals, the imprecise matching between miRNAs and their targets indicates that any given miRNA can bind different mRNAs and may engender an enormous regulatory potential (Ambros, 2008). To disclose the biological events accompanying the alterations of cell invasion caused by NUAK1, online miRNA target prediction databases (miRNA.org (http://www.microrna.org/microrna/searchGenes.do) and Targetscan (http://www.targetscan.org/)) were used to determine the miRNAs that can target NUAK1. The databases predicted miR-204 (Figure 3A), miR-182, miR-96, and miR-708 as the potential miRNAs targeting NUAK1. The four miRNA mimics and negative control (NC) mimics were synthesised (GenePharma Company) and transfected into H1299 and A549 cells (Figure 3B). As shown in Figure 3C, western blot analysis confirmed that the NUAK1 protein levels were indeed reduced drastically in miR-204-transduced cells but pronouncedly elevated in miR-204-silenced cells compared with those in the corresponding control cells. The enhanced miR-182, miR-96, and miR-708 in H1299 cells, however, could not repress NUAK1 expression (Supplementary Figure 3). Based on these results, we hypothesised that NUAK1 was a target of miR-204 in NSCLC, as shown in Figure 3A. Furthermore, RIP analysis following miR-204 transfection demonstrated that mRNAs of NUAK1 could be specifically recruited to the miRNP complex isolated by anti-Ago1 antibody (Figure 3D).
NUAK1 was modulated by miR-204. (A) Predicted binding of miR-204 to 3′UTRs of NUAK1. (B) Relative expression of miR-204 in miR-NC/H1299, miR-204/H1299, miR-204-inhibitor/H1299, miR-NC/A549, miR-204/A549, and miR-204-inhibitor/A549 cells. (C) Western blot analysis of NUAK1 protein expression in miR-NC/H1299, miR-204/H1299, miR-204-inhibitor/H1299, miR-NC/A549, miR-204/A549, and miR-204-inhibitor/A549 cells. β-Actin was used as a loading control. (D) The RIP analysis revealed recruitment of NUAK1 mRNAs to miRNP complex following immunoprecipitation against Ago1. The IgG immunoprecipitation was used as a negative control. (E) MiR-204 and miR-204 mutant oligonucleotides. (F) Luciferase activity of pGL3-NUAK1-3′UTR reporter in indicated cells co-transfected with indicated oligonucleotides. (G) Quantification of indicated cells analysed by transwell invasion assay. *P<0.05 (two-way ANOVA). For (B), (D), (F), and (G), data are presented as mean±s.d. from three independent experiments. Bars, s.d. *P<0.05 (two-way ANOVA).
To further investigate whether the predicted binding site of miR-204 to 3′UTR of NUAK1 is responsible for this regulation, we cloned the 3′UTR of NUAK1 downstream to a luciferase reporter gene (wt-NUAK1). We co-transfected wt-NUAK1 vector and miR-204, miR-204 inhibitor, miR-204 mutant, or negative control into A549 and H1299 cells (Figure 3E). We found that transfection of miR-204 oligonucleotides abrogated the expression of luciferase, and such suppressive effects could be reversed by anti-miR-204 oligonucleotides. However, miR-204-mediated repression of luciferase activity was abolished by the mutant putative binding site (Figure 3F). Taken together, these results indicate that miR-204 downregulates NUAK1 expression by directly targeting its 3′UTR.
Increased expression of miR-204 inhibited NSCLC cell invasion in vitro
To elucidate the functional significance of microRNA-mediated suppression of NUAK1 in cell invasiveness, we re-expressed open reading frames (ORFs, without their respective 3′UTRs) of NUAK1 genes in miR-204-overexpressing cells and assessed cell invasiveness by transwell matrix penetration assay. As expected, restoration of NUAK1 significantly rescued the invasiveness impaired by miR-204 (Figure 3G), suggesting that downregulation of NUAK1 was functionally important for the inhibitory effect of miR-204 on NSCLC cell invasion.
MiR-204 was downregulated in NSCLC
To investigate the potential role of miR-204 in the development and progression of NSCLC, we detected the expression of miR-204 in NSCLC cell lines and tissues. Real-time PCR analysis showed that miR-204 was ubiquitously expressed at lower levels in a series of four human lung cancer cell lines than in BEAS-2B cell line (Figure 4A). In parallel, as shown in Figure 4B, miR-204 expression was markedly decreased in most of the 15 collected frozen NSCLC tumour tissues as compared with that in paired ANTs. These data implied that miR-204 expression was obviously decreased in NSCLC.
Downregulation of miR-204 and its relevance to NUAK1 expression in human NSCLC tissues. (A) Real-time PCR analysis of miR-204 in human bronchial cell line BEAS-2B and human NSCLC cell lines (A549, Calu, SPC-A1, and H1299). (B) Relative expression of miR-204 in 15 pairs of frozen NSCLC tumour tissues (T) and their corresponding adjacent nontumour tissues (ANTs) by real-time PCR analysis. (C) In human NSCLC tissues, miR-204 has a negative corelation with NUAK1 expression. (D) Kaplan–Meier analysis of correlation between miR-204 level and survival of NSCLC patients in stage I–II subgroup. (E) Kaplan–Meier analysis of correlation between miR-204 level and survival of NSCLC patients in stage III–IV subgroup. The P-values were calculated by the log-rank test, and P<0.05 was considered statistically significant.
MiR-204 downregulation has clinical relevance to NUAK1 upregulation in human NSCLC
To examine whether the biological effects of miR-204 downregulation on NSCLC cell invasion were clinically relevant, we examined the correlation of miR-204 expression with the expression of NUAK1 in clinical specimens of NSCLC. As shown in Figure 4C, significantly diminished expression of miR-204 in conjunction with elevated expression of NUAK1 was found in NSCLC lesions, suggesting that the inhibitory effect of miR-204 on NUAK1 was clinically relevant in NSCLC. Moreover, we showed that miR-204 expression was more decreased in NSCLC samples from patients with lymphatic and distant metastasis than those from patients without lymphatic and distant metastasis, and this result is consistent with the forementioned high expression of NUAK1 in metastatic lymph nodes (Supplementary Table 2).
Decreased expression of miR-204 correlates with NSCLC clinicopathologic characteristics and the overall survival of NSCLC patients
The decreased expression of miR-204 in lung cancer tissues and cells prompted us to evaluate the clinical significance of miR-204 in the progression of NSCLC. We examined a cohort of 80 archived paraffin-embedded specimens of NSCLC and 15 selected frozen (liquid nitrogen) NSCLC tissues (Supplementary Table 2). In accordance with aforementioned clinical significance of NUAK1, the data showed that miR-204 was differentially detectable in all NSCLC specimens. The expression of miR-204 was closely correlated with NSCLC clinical staging (P=0.003), lymph node metastasis (P=0.003), and distant metastasis (P=0.006). In addition, Kaplan–Meier analysis indicated that patients with low miR-204 expression in their lung tumours had a shorter median survival time than those with high miR-204 expression in either the stage I–II subgroup (n=47; P=0.044, log-rank; Figure 4D) or the stage III–IV subgroup (n=48; P=0.007, log-rank; Figure 4E). The median survival time of staging I+II subgroup with high miR-204 expression (44.280±2.11 months, 95% confidence interval: 40.144–48.416) was significantly longer than that of patients with low miR-204 expression (35.227±2.681 months, 95% confidence interval: 29.973–40.481). Similarly, the median survival time of staging III+IV subgroup with high miR-204 expression (30.923±4.088 months, 95% confidence interval: 22.910–38.936) was significantly longer than that of patients with low miR-204 expression (19.457±1.598 months, 95% confidence interval: 15.791–22.209). Thus, decreased expression of miR-204 appeared to be a risk factor indicating high invasion and poor prognosis.
MiR-204 inhibited NSCLC tumour invasion by downregulating NUAK1 expression in vivo
To assess the biological functions of NUAK1 and miR-204 in NSCLC in vivo, we employed an experimental model of lung metastasis allowing characterisation of tumour cell extravasation and colonisation in lungs of SCID mice. To initiate the study, 1 million of Scr/H1299, SiNUAK1/H1299, miR-NC/H1299, or miR-204/H1299 cells were transplanted into SCID mice via tail vein injection. As shown in Figure 5A and Supplementary Figure 4A and B, micrometastatic foci were scored after 4 weeks of development in the lung. The number of tumour nodules was less in the lungs of mice injected with SiNUAK1/H1299 and miR-204/H1299 cells than that in the mice injected with Scr/H1299 and miR-NC/H1299 cells. Furthermore, the expression of NUAK1 in the tumour xenograft was lower in the mice injected with SiNUAK1/H1299 and miR-204/H1299 cells than that of the mice injected with Scr/H1299 and miR-NC/H1299 cells (Figure 5B). Thus, our results clearly indicate that NUAK1 can promote metastasis of human NSCLC cells and miR-204 can modulate NUAK1 in vivo. Taken together, these observations indicate that miR-204 can significantly inhibit NUAK1 expression and suppress experimental metastasis of NSCLC.
Downregulation of NUAK1 and upregulation of miR-204 inhibited invasion of human NSCLC in vivo. (A) The numbers of lung metastasis of indicated SCID mice groups (n=10). (B) Western blot analysis of NUAK1 in implanted tumours of the mice injected with Scr/H1299, SiNUAK1/H1299, miR-NC/H1299, and miR-204/H1299 cells. The results were from a representative of at least three repeated experiments.
Downregulation of miR-204 is due to hypermethylation in the promoter region of miR-204 gene
Promoter hypermethylation is thought to be an alternative mechanism to downregulate tumour suppressor genes in human cancers (Egger et al, 2004; Kozaki et al, 2008; Balaguer et al, 2010). To investigate whether the downregulation of miR-204 is due to hypermethylation of the promoter region of miR-204, we analysed the methylation level of miR-204 via bisulphite genomic-sequencing PCR (BSP) in both NSCLC cell lines and tissues. The BSP results showed higher methylation levels of miR-204 in genomic DNA obtained from NSCLC cells and clinical NSCLC tissue samples, and lower methylation levels of miR-204 in BEAS-2B cell line, strongly suggesting an essential role of promoter methylation in miR-204 downregulation (Figure 6A).
Downregulation of miR-204 is due to hypermethylation in the promoter region of miR-204 gene. (A) The methylation levels of miR-204 genomic region in indicated BEAS-2B cells, NSCLC cell lines, and NSCLC specimens. (B) Real-time PCR analysis of miR-204 expression in indicated cells treated with 5 μ M 5-aza-dC or DMSO for 72 h. Data are presented as mean±s.d., *P<0.05. (C) Invasive abilities of H1299 and A549 cells with 5 μ M 5-aza-dC or DMSO treatment for 72 h. Data are presented as mean±s.d., *P<0.05. (D) The numbers of visualised lung metastasis foci of indicated SCID mice groups intravenously injected with 5-aza-dC (400 μ M) or DMSO once a day for 1 week (n=10).
To further validate this finding, we treated the NSCLC cell lines with 5-aza-dC (DNA methyltransferase inhibitor) for 72 h and found that the inhibition of DNA methylation significantly promoted the expression of miR-204 (Figure 6B). We also observed that 5-aza-dC treatment obviously inhibited the invasive ability of H1299 and A549 cells (Figure 6C). In addition, 5-aza-dC treatment significantly suppressed experimental lung metastasis of H1299 NSCLC tumours in SCID mice (Figure 6D). Taken together, these findings strongly suggested that the expression of miR-204 was regulated by promoter methylation in NSCLC cells.
Discussion
In the present study, we first identified the high expression of NUAK1 in both primary NSCLC tissues and cell lines. The NUAK1 is functionally associated with NSCLC metastasis and high expression of NUAK1 is a predictor of poor prognosis for NSCLC patients. At the same time, we found a novel role of NUAK1 in modulating the migration and invasion of NSCLC cells. Downregulation of NUAK1 by siRNA impaired the migration ability of NSCLC cells, and inhibited EGF-induced polymerisation of F-actin and phosphorylation of both cofilin and LIMK1. In addition, downregulation of NUAK1 inhibited MT1-MMP expression and induced decreased activation of MMP-2 and MMP-9 (Li and Tai, 2012; Perlikos et al, 2013). Furthermore, this study suggested that NUAK1 played crucial roles in mTOR pathway.
Meanwhile, this study provides the first evidence that miR-204 plays a role in inhibiting migration and invasion of lung cancer by directly targeting NUAK1, in accordance with the function of miR-211 in melanoma (Bell et al, 2014). By transduction of miR-204 or miR-204 inhibitor into H1299 and A549 cells, we clearly demonstrated that miR-204 can significantly repress NUAK1 expression and play an important role in NSCLC invasion and migration both in vitro and in vivo. Furthermore, overexpression of NUAK1 without 3′UTR led to increased invasion of H1299 cells and significantly reversed the inhibitory effect of miR-204 mimics on the migratory and invasive abilities of H1299 cells. Aside from this, in vitro 3′UTR luciferase assay and RIP analysis also confirmed that miR-204 exerted its effects by targeting NUAK1 via its 3′UTR.
To further verify the relationship between miR-204 and the malignant phenotype of lung cancers, we observed that the expression of miR-204 was apparently low in both NSCLC tissues and tumour cell lines. In addition, inverse correlation between the expression of miR-204 and NUAK1 was evidenced in our clinical relevance and animal experiments study. Particularly, low expression of miR-204 is significantly associated with decreased survival, lymphatic metastasis, and distant metastasis in NSCLC patients. These data are consistent with most of the previous research (Lam et al, 2011; Chung et al, 2012; Gong et al, 2012; Sacconi et al, 2012; Chen et al, 2013; Pignot et al, 2013; Zhang et al, 2013), further suggesting a tumour-suppressive function of miR-204.
Epigenetic modifications have been shown to be crucial mediators underlying the downregulation of miRNA expression and to display a tight correlation with carcinogenesis (Wynter, 2006; Ren et al, 2011). Our data demonstrated that the hypermethylation of the upstream promoter of miR-204 led to the downregulation of miR-204 in NSCLC cell lines and tissues. Moreover, 5-aza-dC, the DNA methyltransferase inhibitor, can increase miR-204 expression in NSCLC cell lines and can reduce the invasive ability of cancer cells. Based on these findings, the methylation status of miR-204 probably acts as a potential biomarker for NSCLC prognosis.
In conclusion, we have shown that NUAK1 is overexpressed in NSCLC and plays important roles in NSCLC invasion. More remarkably, our findings suggest a novel role for miR-204 in inhibiting NUAK1 expression and in suppressing migration and invasion of lung cancer. Furthermore, miR-204 is partly downregulated by hypermethylation of its promoter. These data suggest that expression levels of NUAK1 and miR-204 and the methylation status of miR-204 may serve as potential biomarkers of NSCLC, and miR-204 may have potential therapeutic value in cancer therapy.
Change history
09 December 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN (1987) A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res 47 (12): 3239–3245.
Ambros V (2008) The evolution of our thinking about microRNAs. Nat Med 14 (10): 1036–1040.
Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393 (6687): 805–809.
Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, Goel A (2010) Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res 70 (16): 6609–6618.
Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, Janas MM, Postolsky B, Goldberg MS, Shamir R, Levy C (2014) Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol 134 (2): 441–451.
Buyukcelik A, Yalcin B, Utkan G (2004) Multidisciplinary management of lung cancer. N Engl J Med 350 (19): 2008–2010.
Cai J, Wu J, Zhang H, Fang L, Huang Y, Yang Y, Zhu X, Li R, Li M (2013) miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res 73 (2): 756–766.
Chang XZ, Yu J, Liu HY, Dong RH, Cao XC (2012) ARK5 is associated with the invasive and metastatic potential of human breast cancer cells. J Cancer Res Clin Oncol 138 (2): 247–254.
Chen Z, Sangwan V, Banerjee S, Mackenzie T, Dudeja V, Li X, Wang H, Vickers SM, Saluja AK (2013) miR-204 mediated loss of Myeloid cell leukemia-1 results in pancreatic cancer cell death. Mol Cancer 12 (1): 105.
Cheng GZ, Zhang W, Wang LH (2008) Regulation of cancer cell survival, migration, and invasion by Twist: AKT2 comes to interplay. Cancer Res 68 (4): 957–960.
Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, Barroilhet LM, Ng AS, Wong RR, Wang VW, Mok SC, Smith DI, Berkowitz RS, Wong YF (2012) Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer 130 (5): 1036–1045.
Das G, Shiras A, Shanmuganandam K, Shastry P (2011) Rictor regulates MMP-9 activity and invasion through Raf-1-MEK-ERK signaling pathway in glioma cells. Mol Carcinog 50 (6): 412–423.
Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429 (6990): 457–463.
Garzon R, Calin GA, Croce CM (2009) MicroRNAs in cancer. Annu Rev Med 60: 167–179.
Garzon R, Marcucci G (2012) Potential of microRNAs for cancer diagnostics, prognostication and therapy. Curr Opin Oncol 24 (6): 655–659.
Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS (2004) Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304 (5671): 743–746.
Gong M, Ma J, Li M, Zhou M, Hock JM, Yu X (2012) MicroRNA-204 critically regulates carcinogenesis in malignant peripheral nerve sheath tumors. Neuro Oncol 14 (8): 1007–1017.
Imam JS, Plyler JR, Bansal H, Prajapati S, Bansal S, Rebeles J, Chen HI, Chang YF, Panneerdoss S, Zoghi B, Buddavarapu KC, Broaddus R, Hornsby P, Tomlinson G, Dome J, Vadlamudi RK, Pertsemlidis A, Chen Y, Rao MK (2012) Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One 7 (12): e52397.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61 (2): 69–90.
Khan N, Afaq F, Khusro FH, Mustafa Adhami V, Suh Y, Mukhtar H (2012) Dual inhibition of phosphatidylinositol 3-kinase/Akt and mammalian target of rapamycin signaling in human nonsmall cell lung cancer cells by a dietary flavonoid fisetin. Int J Cancer 130 (7): 1695–1705.
Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J (2008) Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res 68 (7): 2094–2105.
Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37 (5): 495–500.
Kusakai G, Suzuki A, Ogura T, Miyamoto S, Ochiai A, Kaminishi M, Esumi H (2004) ARK5 expression in colorectal cancer and its implications for tumor progression. Am J Pathol 164 (3): 987–995.
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294 (5543): 853–858.
Lam EK, Wang X, Shin VY, Zhang S, Morrison H, Sun J, Ng EK, Yu J, Jin H (2011) A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res 3 (2): 209–218.
Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu Y, Li J, Hasina R, Cheng C, Lingen MW, Gerstein MB, Weichselbaum RR, Xing HR, Lussier YA (2010) Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol 6 (4): e1000730.
Li H, Yin C, Zhang B, Sun Y, Shi L, Liu N, Liang S, Lu S, Liu Y, Zhang J, Li F, Li W, Liu F, Sun L, Qi Y (2013) PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis 34 (9): 2145–2155.
Li J, Guan HY, Gong LY, Song LB, Zhang N, Wu J, Yuan J, Zheng YJ, Huang ZS, Li M (2008) Clinical significance of sphingosine kinase-1 expression in human astrocytomas progression and overall patient survival. Clin Cancer Res 14 (21): 6996–7003.
Li X, Tai HH (2012) Increased expression of matrix metalloproteinases mediates thromboxane A2-induced invasion in lung cancer cells. Curr Cancer Drug Targets 12 (6): 703–715.
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 (7027): 769–773.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435 (7043): 834–838.
Lu S, Niu N, Guo H, Tang J, Guo W, Liu Z, Shi L, Sun T, Zhou F, Li H, Zhang J, Zhang B (2013) ARK5 promotes glioma cell invasion, and its elevated expression is correlated with poor clinical outcome. Eur J Cancer 49 (3): 752–763.
McAuliffe PF, Meric-Bernstam F, Mills GB, Gonzalez-Angulo AM (2010) Deciphering the role of PI3K/Akt/mTOR pathway in breast cancer biology and pathogenesis. Clin Breast Cancer 10 (Suppl 3): S59–S65.
Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K (2005) Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J Cell Biol 171 (2): 349–359.
Perlikos F, Harrington KJ, Syrigos KN (2013) Key molecular mechanisms in lung cancer invasion and metastasis: a comprehensive review. Crit Rev Oncol Hematol 87 (1): 1–11.
Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B, Amsellem-Ouazana D, Bieche I (2013) microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer 132 (11): 2479–2491.
Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, Zhang L (2011) An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ Health Perspect 119 (1): 11–19.
Ryan BM, Robles AI, Harris CC (2010) Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 10 (6): 389–402.
Sacconi A, Biagioni F, Canu V, Mori F, Di Benedetto A, Lorenzon L, Ercolani C, Di Agostino S, Cambria AM, Germoni S, Grasso G, Blandino R, Panebianco V, Ziparo V, Federici O, Muti P, Strano S, Carboni F, Mottolese M, Diodoro M, Pescarmona E, Garofalo A, Blandino G (2012) miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis 3: e423.
Suzuki A, Kusakai G, Kishimoto A, Lu J, Ogura T, Esumi H (2003) ARK5 suppresses the cell death induced by nutrient starvation and death receptors via inhibition of caspase 8 activation, but not by chemotherapeutic agents or UV irradiation. Oncogene 22 (40): 6177–6182.
Suzuki A, Lu J, Kusakai G, Kishimoto A, Ogura T, Esumi H (2004) ARK5 is a tumor invasion-associated factor downstream of Akt signaling. Mol Cell Biol 24 (8): 3526–3535.
Takemoto N, Tada M, Hida Y, Asano T, Cheng S, Kuramae T, Hamada J, Miyamoto M, Kondo S, Moriuchi T (2007) Low expression of reversion-inducing cysteine-rich protein with Kazal motifs (RECK) indicates a shorter survival after resection in patients with adenocarcinoma of the lung. Lung Cancer 58 (3): 376–383.
Tan LP, Seinen E, Duns G, de Jong D, Sibon OC, Poppema S, Kroesen BJ, Kok K, van den Berg A (2009) A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res 37 (20): e137.
Turner DP, Findlay VJ, Moussa O, Semenchenko VI, Watson PM, LaRue AC, Desouki MM, Fraig M, Watson DK (2011) Mechanisms and functional consequences of PDEF protein expression loss during prostate cancer progression. Prostate 71 (16): 1723–1735.
Ventura A, Jacks T (2009) MicroRNAs and cancer: short RNAs go a long way. Cell 136 (4): 586–591.
Ventura AC, Jackson TL, Merajver SD (2009) On the role of cell signaling models in cancer research. Cancer Res 69 (2): 400–402.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103 (7): 2257–2261.
Weng Y, Cai M, Zhu J, Geng J, Zhu K, Jin X, Ding W (2013) Matrix metalloproteinase activity in early-stage lung cancer. Onkologie 36 (5): 256–259.
Wynter CV (2006) The dialectics of cancer: a theory of the initiation and development of cancer through errors in RNAi. Med Hypotheses 66 (3): 612–635.
Xu Z, Liu D, Fan C, Luan L, Zhang X, Wang E (2013) DIXDC1 increases the invasion and migration ability of non-small-cell lung cancer cells via the PI3K-AKT/AP-1 pathway. Mol Carcinog 53 (11): 917–925.
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M (2013) Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res 73 (2): 990–999.
Yoo CB, Jones PA (2006) Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 5 (1): 37–50.
Zhang B, Gu F, She C, Guo H, Li W, Niu R, Fu L, Zhang N, Ma Y (2009a) Reduction of Akt2 inhibits migration and invasion of glioma cells. Int J Cancer 125 (3): 585–595.
Zhang B, Ma Y, Guo H, Sun B, Niu R, Ying G, Zhang N (2009b) Akt2 is required for macrophage chemotaxis. Eur J Immunol 39 (3): 894–901.
Zhang F, Zhang X, Li M, Chen P, Zhang B, Guo H, Cao W, Wei X, Cao X, Hao X, Zhang N (2010) mTOR complex component Rictor interacts with PKCzeta and regulates cancer cell metastasis. Cancer Res 70 (22): 9360–9370.
Zhang L, Wang X, Chen P (2013) MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer 13: 290.
Acknowledgements
This work was supported by National Natural Scientific Foundation of China (81072068, 81472365), The Young and Middle-Aged Scientists Research Awards Foundation of Shandong Province (2010BSB14050), Foundation of Shandong Educational Committee (J14LK13), Scientific Foundation of Shandong Province (ZR2011HL047), Shandong Province Outstanding Youth Scientist Foundation Plan (BS2013YY020), Shandong Medical and Health Science and Technology Development Plan (2013WS0282) and Foundation of Weifang Scientific Committee (20121230).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Shi, L., Zhang, B., Sun, X. et al. MiR-204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer 111, 2316–2327 (2014). https://doi.org/10.1038/bjc.2014.580
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.580
Keywords
This article is cited by
-
miR-204 suppresses uveal melanoma cell migration and invasion through negative regulation of RAB22A
Functional & Integrative Genomics (2023)
-
MicroRNA-1182 and let-7a exert synergistic inhibition on invasion, migration and autophagy of cholangiocarcinoma cells through down-regulation of NUAK1
Cancer Cell International (2021)
-
MiR-206 may suppress non-small lung cancer metastasis by targeting CORO1C
Cellular & Molecular Biology Letters (2020)
-
miR-622 is a novel potential biomarker of breast carcinoma and impairs motility of breast cancer cells through targeting NUAK1 kinase
British Journal of Cancer (2020)
-
MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2
Cellular & Molecular Biology Letters (2019)