Article
|
Open Access
Featured
-
-
Article
| Open Access Relaxed targeting rules help PIWI proteins silence transposons
Relaxed targeting rules help PIWI proteins silence transposons
Of the two types of Argonaute proteins produced by animals, AGO and PIWI, PIWI proteins can bind RNAs with less complementarity, enabling efficient silencing of transposons without the need to produce new RNA guides.
- Ildar Gainetdinov
- , Joel Vega-Badillo
- & Phillip D. Zamore
-
Article |
 Sequence determinant of small RNA production by DICER
Sequence determinant of small RNA production by DICER
Massively parallel assays reveal a highly conserved sequence motif termed the GYM motif, which potentiates RNA interference by directing Dicer-mediated small RNA processing.
- Young-Yoon Lee
- , Haedong Kim
- & V. Narry Kim
-
Article
| Open Access GTSF1 accelerates target RNA cleavage by PIWI-clade Argonaute proteins
GTSF1 accelerates target RNA cleavage by PIWI-clade Argonaute proteins
The evolutionarily conserved RNA-binding protein GTSF1 and its homologues interact with members of the PIWI class of Argonaute proteins, increasing the efficiency of the RNA-cleaving activity of PIWI proteins, an essential function across the animal kingdom.
- Amena Arif
- , Shannon Bailey
- & Phillip D. Zamore
-
Article
| Open Access Structural insights into dsRNA processing by Drosophila Dicer-2–Loqs-PD
Structural insights into dsRNA processing by Drosophila Dicer-2–Loqs-PD
Structures of the Dcr-2–Loqs-PD complex while it is processing a double-stranded RNA (dsRNA) substrate elucidate the interactions between Dcr-2 and Loqs-PD, and show that Dcr-2 undergoes substantial conformational changes during a dsRNA-processing cycle.
- Shichen Su
- , Jia Wang
- & Jinbiao Ma
-
Article
| Open Access Structure of the Dicer-2–R2D2 heterodimer bound to a small RNA duplex
Structure of the Dicer-2–R2D2 heterodimer bound to a small RNA duplex
Cryo-electron microscopy structures of Drosophila Dicer-2–R2D2 complexes with and without small interfering RNA reveal how the RNA is presented to Argonaute in the correct orientation for viral gene silencing.
- Sonomi Yamaguchi
- , Masahiro Naganuma
- & Osamu Nureki
-
Article |
 Structure and function of virion RNA polymerase of a crAss-like phage
Structure and function of virion RNA polymerase of a crAss-like phage
The RNA polymerase from the crAss-like bacteriophage phi14:2, which is translocated into the host cell with phage DNA and transcribes early phage genes, is structurally most similar to eukaryotic RNA interference polymerases, suggesting that the latter have a phage origin.
- Arina V. Drobysheva
- , Sofia A. Panafidina
- & Maria L. Sokolova
-
Article |
 C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance
C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance
Exposing Caenorhabditis elegans to non-coding small RNAs from pathogenic Pseudomonas aeruginosa induces avoidance behaviours in treated worms and their progeny, which reveals how C. elegans discriminates between bacterial species in its microbial environment.
- Rachel Kaletsky
- , Rebecca S. Moore
- & Coleen T. Murphy
-
Article |
 DNA targeting and interference by a bacterial Argonaute nuclease
DNA targeting and interference by a bacterial Argonaute nuclease
Argonaute protein from the bacterium C. butyricum targets multicopy genetic elements and functions in the suppression of plasmid and phage propagation, and there appears to be a DNA-mediated immunity pathway in prokaryotes.
- Anton Kuzmenko
- , Anastasiya Oguienko
- & Andrey Kulbachinskiy
-
Article |
 SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation
SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation
Newly identified protein SPOCD1 is crucial in de novo DNA methylation directed by PIWI proteins and piRNAs, helping to control DNA silencing in mouse male germline.
- Ansgar Zoch
- , Tania Auchynnikava
- & Dónal O’Carroll
-
Article |
 Base-pair conformational switch modulates miR-34a targeting of Sirt1 mRNA
Base-pair conformational switch modulates miR-34a targeting of Sirt1 mRNA
Repression of a messenger RNA by a cognate microRNA depends not only on complementary base pairing, but also on the rearrangement of a single base pair, producing a conformation that fits better within the human Ago2 protein.
- Lorenzo Baronti
- , Ileana Guzzetti
- & Katja Petzold
-
Article |
 poly(UG)-tailed RNAs in genome protection and epigenetic inheritance
poly(UG)-tailed RNAs in genome protection and epigenetic inheritance
In Caenorhabditis elegans, the ribonucleotidyltransferase RDE-3 adds alternating uridine and guanosine ribonucleotides to the 3′ termini of RNAs, a key step in RNA interference and thus epigenetic inheritance in the C. elegans germline.
- Aditi Shukla
- , Jenny Yan
- & Scott Kennedy
-
Letter |
 Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation
Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation
In Schizosaccharomyces pombe, histone H3K9 methylation acts synergistically with short interfering RNA to perpetuate gene silencing during multiple mitotic and meiotic cell divisions.
- Ruby Yu
- , Xiaoyi Wang
- & Danesh Moazed
-
Article |
 Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance
Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance
ZNFX-1 and WAGO-4 localize to germ granules in early Caenorhabditis elegans embryogenesis and later separate to form independent liquid-like droplets, and the temporal and spatial ordering of these droplets may help cells to organize complex RNA processing pathways.
- Gang Wan
- , Brandon D. Fields
- & Scott Kennedy
-
Article |
 SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production
SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production
The chromatin remodelling protein CHR2 interacts with Serrate in Arabidopsis to regulate microRNA biogenesis.
- Zhiye Wang
- , Zeyang Ma
- & Xiuren Zhang
-
Letter |
 Transcription elongation factors represent in vivo cancer dependencies in glioblastoma
Transcription elongation factors represent in vivo cancer dependencies in glioblastoma
An in vivo RNA interference screening strategy in glioblastoma enabled the identification of a host of epigenetic targets required for glioblastoma cell survival that were not identified by parallel standard screening in cell culture, including the transcription pause–release factor JMJD6, and could be a powerful tool to uncover new therapeutic targets in cancer.
- Tyler E. Miller
- , Brian B. Liau
- & Jeremy N. Rich
-
Letter |
 Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription
Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription
Heterochromatin formation involves histone H3 methylation, with H3K9me2 defining a distinct heterochromatin state that is transcriptionally permissive and can couple with RNAi, and the transition to non-permissive H3K9me3 required for the epigenetic heritability of heterochromatin.
- Gloria Jih
- , Nahid Iglesias
- & Danesh Moazed
-
Article |
 An Argonaute phosphorylation cycle promotes microRNA-mediated silencing
An Argonaute phosphorylation cycle promotes microRNA-mediated silencing
The application of genome-wide CRISPR–Cas9 screening coupled with a fluorescent reporter to interrogate the microRNA pathway reveals that continual transient phosphorylation of Argonaute 2 is required to maintain the global efficiency of microRNA-mediated repression.
- Ryan J. Golden
- , Beibei Chen
- & Joshua T. Mendell
-
Letter |
 Genetic and mechanistic diversity of piRNA 3′-end formation
Genetic and mechanistic diversity of piRNA 3′-end formation
Drosophila have two pathways for PIWI-interacting RNA (piRNA) 3′-end formation—depending on which pathway is used, piRNA biogenesis is directed towards either cytoplasmic or nuclear PIWI protein effectors, which balances post-transcriptional versus transcriptional transposon silencing.
- Rippei Hayashi
- , Jakob Schnabl
- & Julius Brennecke
-
Letter |
 Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm
Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm
Maternal mRNAs are tethered within the Drosophila germ plasm via base-pairing interactions between mRNAs and piRNPs containing the Aub Piwi protein; the preference for certain mRNAs to be tethered appears to be related to their longer length, which provides more potential piRNP-binding sites, and the results suggest a new role for piRNAs in germ-cell specification independent of their role in transposon silencing.
- Anastassios Vourekas
- , Panagiotis Alexiou
- & Zissimos Mourelatos
-
Letter |
 The spliceosome is a therapeutic vulnerability in MYC-driven cancer
The spliceosome is a therapeutic vulnerability in MYC-driven cancer
Splicing factors such as BUD31 are identified in a synthetic-lethal screen with cells overexpressing the transcription factor MYC; oncogenic MYC leads to an increase in pre-mRNA synthesis, and spliceosome inhibition impairs the growth and tumorigenicity of MYC-dependent breast cancers, suggesting that spliceosome components may be potential therapeutic targets for MYC-driven cancers.
- Tiffany Y.-T. Hsu
- , Lukas M. Simon
- & Thomas F. Westbrook
-
Letter |
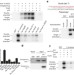 Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex
Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex
The assembly of single Drosophila RNA-induced silencing complexes (RISCs) is reconstituted using seven purified proteins, revealing that chaperones help stabilize the interaction of the protein heterodimer Dicer-2–R2D2 bound to the short interfering RNA with Ago2.
- Shintaro Iwasaki
- , Hiroshi M. Sasaki
- & Yukihide Tomari
-
Letter |
 The Paf1 complex represses small-RNA-mediated epigenetic gene silencing
The Paf1 complex represses small-RNA-mediated epigenetic gene silencing
The fission yeast is shown to have a mechanism to prevent small RNAs from inducing heterochromatin and epigenetic gene silencing; this protective model involves the highly conserved Paf1 complex, which is known to promote transcription and processing of pre-mRNA, and protects protein-coding genes from unwanted silencing by spurious transcripts.
- Katarzyna Maria Kowalik
- , Yukiko Shimada
- & Marc Bühler
-
Letter |
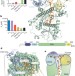 Mechanism of Dis3l2 substrate recognition in the Lin28–let-7 pathway
Mechanism of Dis3l2 substrate recognition in the Lin28–let-7 pathway
The structure of mouse Dis3l2 bound to an oligoU substrate shows a funnel-like substrate-binding site with the RNA being fed into the active site along a path that is distinct from that seen in the related catalytic subunit of the exosome — 12 uracils of the oligoU-tailed RNA are recognized in a complex network of interactions, suggesting the basis for target specificity.
- Christopher R. Faehnle
- , Jack Walleshauser
- & Leemor Joshua-Tor
-
Letter |
 Antifungal drug resistance evoked via RNAi-dependent epimutations
Antifungal drug resistance evoked via RNAi-dependent epimutations
The human fungal pathogen Mucor circinelloides develops spontaneous resistance to an antifungal drug both through mutation and through a newly identified epigenetic RNA-mediated pathway; RNA interference is spontaneously triggered to silence the fkbA gene, giving rise to drug-resistant epimutants that revert to being drug-sensitive once again when grown in the absence of drug.
- Silvia Calo
- , Cecelia Shertz-Wall
- & Joseph Heitman
-
Article |
 Genome-defence small RNAs exapted for epigenetic mating-type inheritance
Genome-defence small RNAs exapted for epigenetic mating-type inheritance
The molecular basis for mating-type determination in the ciliate Paramecium has been elucidated, revealing a novel function for a class of small RNAs — these scnRNAs are typically involved in reprogramming the Paramecium genome during sexual reproduction by recognizing and excising transposable elements, but they are now found to be co-opted to switch off expression of the newly identified mating-type gene mtA by excising its promoter, and to mediate epigenetic inheritance of mating types across sexual generations.
- Deepankar Pratap Singh
- , Baptiste Saudemont
- & Eric Meyer
-
Letter |
 PTEN action in leukaemia dictated by the tissue microenvironment
PTEN action in leukaemia dictated by the tissue microenvironment
A mouse model of T-cell leukaemia is used to test whether PTEN loss is required for tumour maintenance as well as initiation; although it had little effect on tumour load in haematopoietic organs, PTEN reactivation reduced the CCR9-dependent tumour dissemination to the intestine that was amplified on PTEN loss, exposing the importance of tumour microenvironment in PTEN-deficient settings.
- Cornelius Miething
- , Claudio Scuoppo
- & Scott W. Lowe
-
Letter |
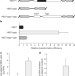 DNA-guided DNA interference by a prokaryotic Argonaute
DNA-guided DNA interference by a prokaryotic Argonaute
Here, Argonaute from the prokaryote Thermus thermophilus is shown to use small DNA guides to interfere directly with invading foreign DNA, rather than being involved in RNA-guided RNA interference, as observed in eukaryotes.
- Daan C. Swarts
- , Matthijs M. Jore
- & John van der Oost
-
Article |
 RNAi screens in mice identify physiological regulators of oncogenic growth
RNAi screens in mice identify physiological regulators of oncogenic growth
Here, the first genome-wide in vivo RNA interference screens in a mammalian animal model are reported: genes involved in normal and abnormal epithelial cell growth are studied in developing skin tissue in mouse embryos, and among the findings, β-catenin is shown to act as an antagonist to normal epithelial cell growth as well as promoting oncogene-driven growth.
- Slobodan Beronja
- , Peter Janki
- & Elaine Fuchs
-
Letter |
 EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2
EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2
Epidermal growth factor receptor, the product of a human oncogene, suppresses the maturation of specific tumour-suppressor-like microRNAs in response to hypoxic stress through phosphorylation of argonaute 2.
- Jia Shen
- , Weiya Xia
- & Mien-Chie Hung
-
Letter |
 A CRISPR/Cas system mediates bacterial innate immune evasion and virulence
A CRISPR/Cas system mediates bacterial innate immune evasion and virulence
The CRISPR/Cas system known to aid bacterial defences by targeting invading DNA can also act to evade eukaryotic defences through a different class of small RNAs downregulating an endogenous immunogenic bacterial lipoprotein.
- Timothy R. Sampson
- , Sunil D. Saroj
- & David S. Weiss
-
Letter |
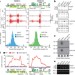 RNAi triggered by specialized machinery silences developmental genes and retrotransposons
RNAi triggered by specialized machinery silences developmental genes and retrotransposons
In the fission yeast Schizosaccharomyces pombe RNA interference (RNAi) machinery promotes heterochromatin assembly and silencing of centromeric repeats; here it is shown that RNAi participates in silencing other genomic regions, such as sexual differentiation genes and retrotransposons, and this process is regulated by developmental and environmental signals.
- Soichiro Yamanaka
- , Sameet Mehta
- & Shiv I. S. Grewal
-
News |
 Animals engineered with pinpoint accuracy
Animals engineered with pinpoint accuracy
More accurate genetic modification has created allergen-free cow's milk and pigs that could serve as a model for atherosclerosis.
- Amy Maxmen
-
Letter |
 A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality
A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality
Double-stranded RNA interference (RNAi) in Caenorhabditis elegans is heritable; here a genetic screen for factors required for RNAi inheritance identifies the nuclear-localized Argonaute gene hrde-1, which acts in the germ cells of progeny to promote multigenerational inheritance of silencing and, also, germline immortality.
- Bethany A. Buckley
- , Kirk B. Burkhart
- & Scott Kennedy
-
Article |
 Structure of yeast Argonaute with guide RNA
Structure of yeast Argonaute with guide RNA
Argonaute proteins are an essential part of the guide-RNA–protein complex that carries out RNA-induced gene silencing; structure–function studies of the yeast complex reveal conserved features of the eukaryotic complex, which underlie formation of the catalytically active conformation.
- Kotaro Nakanishi
- , David E. Weinberg
- & Dinshaw J. Patel
-
Letter |
 An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration
An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration
The transcription factor TBX1 has a role in stem cell activation and self-renewal during long-term tissue regeneration.
- Ting Chen
- , Evan Heller
- & Elaine Fuchs
-
News & Views |
 Genomics decodes drug action
Genomics decodes drug action
Drugs used to treat African sleeping sickness are outdated, and how they enter cells and exert biological effects is poorly understood. A genome-wide study using RNA interference provides valuable insight. See Letter p.232
- Alan H. Fairlamb
-
Letter |
 High-throughput decoding of antitrypanosomal drug efficacy and resistance
High-throughput decoding of antitrypanosomal drug efficacy and resistance
Five current human African trypanosomiasis drugs are used for genome-scale RNA interference target sequencing screens in Trypanosoma brucei, and reveal the transporters, organelles, enzymes and metabolic pathways that function to facilitate antitrypanosomal drug action.
- Sam Alsford
- , Sabine Eckert
- & David Horn
-
News |
 Video animation: RNA interference
Video animation: RNA interference
A video explaining RNA interference from Nature Reviews Genetics.
-
Letter |
 Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila
Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila
- Filippo M. Cernilogar
- , Maria Cristina Onorati
- & Valerio Orlando
-
Letter |
 RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II
RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II
- Mikel Zaratiegui
- , Stephane E. Castel
- & Robert A. Martienssen
-
Letter |
 RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia
RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia
- Johannes Zuber
- , Junwei Shi
- & Christopher R. Vakoc
-
Article |
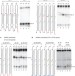 Dicer recognizes the 5′ end of RNA for efficient and accurate processing
Dicer recognizes the 5′ end of RNA for efficient and accurate processing
- Jong-Eun Park
- , Inha Heo
- & V. Narry Kim
-
Letter |
 Non-apoptotic role of BID in inflammation and innate immunity
Non-apoptotic role of BID in inflammation and innate immunity
- Garabet Yeretssian
- , Ricardo G. Correa
- & Maya Saleh
-
News |
 Drug giants turn their backs on RNA interference
Drug giants turn their backs on RNA interference
A once much-touted technique faces a difficult transition to the clinic.
- Heidi Ledford
-
Letter |
 A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity
A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity
Realizing the full potential of human embryonic stem cells (hESCs) in research and clinical applications requires a detailed understanding of the genetic network that governs their unique properties. A genome-wide RNA interference screen identifies a wealth of new regulators of self-renewal and pluripotency properties in hESCs. The transcription factor PRDM14, for example, is required for the maintenance of hESC identity and reprogramming of somatic cells to pluripotency.
- Na-Yu Chia
- , Yun-Shen Chan
- & Huck-Hui Ng
-
Article |
 MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake
MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake
The uptake of calcium by mitochondria has a central role in cell physiology, and an imbalance can trigger cell death. Now the first protein that is localized to the mitochondrion and is specifically required for calcium uptake has been identified. This protein, mitochondrial calcium uptake 1 (MICU1), represents the founding member of a set of proteins required for high-capacity calcium uptake. Its discovery should aid in the full molecular characterization of the mitochondrial calcium uptake pathways.
- Fabiana Perocchi
- , Vishal M. Gohil
- & Vamsi K. Mootha
-
Research Highlights |
Drug development: Virus knockdown
-
Technology Feature |
 From tools to therapies
From tools to therapies
-
Technology Feature |
Table of suppliers

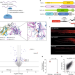 piRNA processing by a trimeric Schlafen-domain nuclease
piRNA processing by a trimeric Schlafen-domain nuclease