Abstract
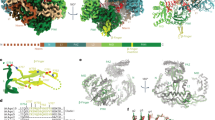
The RNA-induced silencing complex, comprising Argonaute and guide RNA, mediates RNA interference. Here we report the 3.2 Å crystal structure of Kluyveromyces polysporus Argonaute (KpAGO) fortuitously complexed with guide RNA originating from small-RNA duplexes autonomously loaded and processed by recombinant KpAGO. Despite their diverse sequences, guide-RNA nucleotides 1–8 are positioned similarly, with sequence-independent contacts to bases, phosphates and 2′-hydroxyl groups pre-organizing the backbone of nucleotides 2–8 in a near-A-form conformation. Compared with prokaryotic Argonautes, KpAGO has numerous surface-exposed insertion segments, with a cluster of conserved insertions repositioning the N domain to enable full propagation of guide–target pairing. Compared with Argonautes in inactive conformations, KpAGO has a hydrogen-bond network that stabilizes an expanded and repositioned loop, which inserts an invariant glutamate into the catalytic pocket. Mutation analyses and analogies to ribonuclease H indicate that insertion of this glutamate finger completes a universally conserved catalytic tetrad, thereby activating Argonaute for RNA cleavage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Protein Data Bank
Data deposits
The structural coordinates of KpAGO have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) under accession code 4F1N. RNA-sequencing data have been deposited in the Gene Expression Omnibus (http:// www.ncbi.nlm.nih.gov/geo) under accession number GSE37725.
Change history
22 June 2012
The HTML published online on 18 May was the non-final version of the proof; this has now been corrected.
References
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004)
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349 (2004)
Malone, C. D. & Hannon, G. J. Small RNAs as guardians of the genome. Cell 136, 656–668 (2009)
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001)
Elbashir, S. M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001)
Hutvágner, G. et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 (2001)
Matranga, C., Tomari, Y., Shin, C., Bartel, D. P. & Zamore, P. D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 (2005)
Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H. & Siomi, M. C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19, 2837–2848 (2005)
Rand, T. A., Petersen, S., Du, F. & Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629 (2005)
Tomari, Y. & Zamore, P. D. Perspective: machines for RNAi. Genes Dev. 19, 517–529 (2005)
Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009)
Song, J. J., Smith, S. K., Hannon, G. J. & Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 (2004)
Yuan, Y. R. et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19, 405–419 (2005)
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004)
Parker, J. S., Roe, S. M. & Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 23, 4727–4737 (2004)
Ma, J. B. et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670 (2005)
Wang, Y., Sheng, G., Juranek, S., Tuschl, T. & Patel, D. J. Structure of the guide-strand-containing argonaute silencing complex. Nature 456, 209–213 (2008)
Wang, Y. et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926 (2008)
Wang, Y. et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461, 754–761 (2009)
Parker, J. S. How to slice: snapshots of Argonaute in action. Silence 1, 3 (2010)
Makarova, K. S., Wolf, Y. I., van der Oost, J. & Koonin, E. V. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4, 29 (2009)
Meister, G. et al. Identification of novel argonaute-associated proteins. Curr. Biol. 15, 2149–2155 (2005)
Lingel, A., Simon, B., Izaurralde, E. & Sattler, M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426, 465–469 (2003)
Yan, K. S. et al. Structure and conserved RNA binding of the PAZ domain. Nature 426, 468–474 (2003)
Ma, J. B., Ye, K. & Patel, D. J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322 (2004)
Boland, A., Tritschler, F., Heimstadt, S., Izaurralde, E. & Weichenrieder, O. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 11, 522–527 (2010)
Frank, F., Sonenberg, N. & Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822 (2010)
Boland, A., Huntzinger, E., Schmidt, S., Izaurralde, E. & Weichenrieder, O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl Acad. Sci. USA 108, 10466–10471 (2011)
Drinnenberg, I. A. et al. RNAi in budding yeast. Science 326, 544–550 (2009)
Weinberg, D. E., Nakanishi, K., Patel, D. J. & Bartel, D. P. The inside-out mechanism of Dicers from budding yeasts. Cell 146, 262–276 (2011)
Rivas, F. V. et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Struct. Mol. Biol. 12, 340–349 (2005)
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P. & Axtell, M. J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762 (2008)
German, M. A. et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nature Biotechnol. 26, 941–946 (2008)
Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888 (2001)
Haley, B. & Zamore, P. D. Kinetic analysis of the RNAi enzyme complex. Nature Struct. Mol. Biol. 11, 599–606 (2004)
Martinez, J. & Tuschl, T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 18, 975–980 (2004)
Ameres, S. L., Martinez, J. & Schroeder, R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130, 101–112 (2007)
Förstemann, K., Horwich, M. D., Wee, L., Tomari, Y. & Zamore, P. D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130, 287–297 (2007)
Frazão, C. et al. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 443, 110–114 (2006)
Parker, J. S., Roe, S. M. & Barford, D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434, 663–666 (2005)
Pan, Y. & MacKerell, A. D., Jr Altered structural fluctuations in duplex RNA versus DNA: a conformational switch involving base pair opening. Nucleic Acids Res. 31, 7131–7140 (2003)
Schwarz, D. S., Hutvágner, G., Haley, B. & Zamore, P. D. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10, 537–548 (2002)
Chiu, Y. L. & Rana, T. M. siRNA function in RNAi: a chemical modification analysis. RNA 9, 1034–1048 (2003)
Parker, J. S., Parizotto, E. A., Wang, M., Roe, S. M. & Barford, D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell 33, 204–214 (2009)
Mallory, A. C. et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364 (2004)
Nowotny, M., Gaidamakov, S. A., Crouch, R. J. & Yang, W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 (2005)
Hall, T. M. Structure and function of argonaute proteins. Structure 13, 1403–1408 (2005)
Nowotny, M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 10, 144–151 (2009)
Schirle, N. T. & Macrae, I. J. The crystal structure of human Argonaute2. Sciencehttp://dx.doi.org/10.1126/science.1221551 (26 April 2012)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276, 307–326 (1997)
Pape, T. & Schneider, T. R. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 (2004)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 50, 760–763 (1994)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 58, 1948–1954 (2002)
Vagin, A. & Teplyakov, A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. 56, 1622–1624 (2000)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 54, 905–921 (1998)
DeLano, W. L. & Lam, J. W. PyMOL: A communications tool for computational models. Abstracts of Papers of the American Chemical Society 230, (2005)
Grimson, A. et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455, 1193–1197 (2008)
Jeong, H. et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 394, 644–652 (2009)
Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 31–34 (2007)
Mumberg, D., Muller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995)
Sikorski, R. S. & Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics 122, 19–27 (1989)
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14, 953–961 (1998)
Acknowledgements
We thank K. Rajashankar for data processing and phasing, V. Auyeung and D. Shechner for discussions, the Whitehead Genome Technology Core for high-throughput sequencing, and the NE-CAT beamline at the Advanced Photon Source. This work was supported by National Institutes of Health grants AI068776 (D.J.P.) and GM61835 (D.P.B.), a Human Frontier Science Program Long-term Fellowship (K.N.), a fellowship from the Japan Society for the Promotion of Science for Research Abroad (K.N.), and a National Science Foundation graduate research fellowship (D.E.W.). D.P.B. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
All authors designed the study and wrote the manuscript. Structural experiments were performed by K.N. under the supervision of D.J.P. Biochemical experiments were performed by D.E.W. under the supervision of D.P.B.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-6 and Supplementary Figures 1-14. This file has been amended and was replaced on 20 June 2012. (PDF 5212 kb)
Rights and permissions
About this article
Cite this article
Nakanishi, K., Weinberg, D., Bartel, D. et al. Structure of yeast Argonaute with guide RNA. Nature 486, 368–374 (2012). https://doi.org/10.1038/nature11211
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11211
This article is cited by
-
Structural basis for sequence-specific recognition of guide and target strands by the Archaeoglobus fulgidus Argonaute protein
Scientific Reports (2023)
-
Oligomerization-mediated activation of a short prokaryotic Argonaute
Nature (2023)
-
Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex
Nature Communications (2021)
-
Does the Global Outbreak of COVID-19 or Other Viral Diseases Threaten the Stem Cell Reservoir Inside the Body?
Stem Cell Reviews and Reports (2021)
-
Argonaute proteins: structures and their endonuclease activity
Molecular Biology Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.