Featured
-
-
Article |
 Two independent modes of chromatin organization revealed by cohesin removal
Two independent modes of chromatin organization revealed by cohesin removal
Depletion of chromosome-associated cohesin leads to loss of topologically associating domains in interphase chromosomes, without affecting segregation into compartments, and instead, it unmasks a finer compartment structure that reflects local chromatin and transcriptional activity.
- Wibke Schwarzer
- , Nezar Abdennur
- & Francois Spitz
-
Letter |
 Polycomb-like proteins link the PRC2 complex to CpG islands
Polycomb-like proteins link the PRC2 complex to CpG islands
Crystal structures of the Polycomb-like proteins PHF1 and MTF2 with bound DNA and histone peptides show that extended homologous regions of the two proteins form a winged-helix structure that has an unexpected mechanism of binding to unmethylated CpG-containing DNA motifs.
- Haojie Li
- , Robert Liefke
- & Zhanxin Wang
-
Article |
 A heterochromatin-dependent transcription machinery drives piRNA expression
A heterochromatin-dependent transcription machinery drives piRNA expression
Transcription of Drosophila PIWI-interacting RNA (piRNA) clusters is enforced through RNA polymerase II pre-initiation complex formation within repressive heterochromatin, accomplished through the transcription factor IIA subunit paralogue Moonshiner.
- Peter Refsing Andersen
- , Laszlo Tirian
- & Julius Brennecke
-
Brief Communications Arising |
 Elsässer et al. reply
Elsässer et al. reply
- Simon J. Elsässer
- , Kyung-Min Noh
- & Laura A. Banaszynski
-
Brief Communications Arising |
 On the role of H3.3 in retroviral silencing
On the role of H3.3 in retroviral silencing
- Gernot Wolf
- , Rita Rebollo
- & Todd S. Macfarlan
-
Letter |
 ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference
ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference
A high-throughput approach using a DNA-barcoded nucleosome library shows that ISWI chromatin remodellers can distinguish between differently modified nucleosomes.
- Geoffrey P. Dann
- , Glen P. Liszczak
- & Tom W. Muir
-
Letter |
 Allelic reprogramming of 3D chromatin architecture during early mammalian development
Allelic reprogramming of 3D chromatin architecture during early mammalian development
A low-input Hi-C method is used to show that chromatin organization is markedly relaxed in pre-implantation mouse embryos after fertilization and that the subsequent maturation of 3D chromatin architecture is surprisingly slow.
- Zhenhai Du
- , Hui Zheng
- & Wei Xie
-
Letter |
 Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription
Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription
Heterochromatin formation involves histone H3 methylation, with H3K9me2 defining a distinct heterochromatin state that is transcriptionally permissive and can couple with RNAi, and the transition to non-permissive H3K9me3 required for the epigenetic heritability of heterochromatin.
- Gloria Jih
- , Nahid Iglesias
- & Danesh Moazed
-
Letter |
 Phase separation drives heterochromatin domain formation
Phase separation drives heterochromatin domain formation
HP1a can nucleate into foci that display liquid properties during the early stages of heterochromatin domain formation in Drosophila embryos, suggesting that the repressive action of heterochromatin may be mediated in part by emergent properties of phase separation.
- Amy R. Strom
- , Alexander V. Emelyanov
- & Gary H. Karpen
-
Letter |
 Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin
Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin
Phosphorylation or DNA binding promotes the physical partitioning of HP1α out of a soluble aqueous phase into droplets, suggesting that the repressive action of heterochromatin may in part be mediated by the phase separation of HP1.
- Adam G. Larson
- , Daniel Elnatan
- & Geeta J. Narlikar
-
Article |
 Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory
Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory
The metabolic enzyme acetyl coenzyme A synthetase directly regulates gene expression during memory formation by binding to specific genes and providing acetyl coenzyme A for histone acetylation.
- Philipp Mews
- , Greg Donahue
- & Shelley L. Berger
-
Article |
 Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure
Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure
The cryo-electron microscopy structure of Saccharomyces cerevisiae Snf2 chromatin remodeller bound to a nucleosome and a proposed mechanism for DNA translocation by Snf2 are presented.
- Xiaoyu Liu
- , Meijing Li
- & Zhucheng Chen
-
Letter |
 Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition
Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition
Using a single-nucleus Hi-C protocol, the authors find that spatial organization of chromatin during oocyte-to-zygote transition differs between paternal and maternal nuclei within a single-cell zygote.
- Ilya M. Flyamer
- , Johanna Gassler
- & Kikuë Tachibana-Konwalski
-
Article |
 3D structures of individual mammalian genomes studied by single-cell Hi-C
3D structures of individual mammalian genomes studied by single-cell Hi-C
A chromosome conformation capture method in which single cells are first imaged and then processed enables intact genome folding to be studied at a scale of 100 kb, validated, and analysed to generate hypotheses about 3D genomic interactions and organisation.
- Tim J. Stevens
- , David Lando
- & Ernest D. Laue
-
Article |
 Complex multi-enhancer contacts captured by genome architecture mapping
Complex multi-enhancer contacts captured by genome architecture mapping
A technique called genome architecture mapping (GAM) involves sequencing DNA from a large number of thin nuclear cryosections to develop a map of genome organization without the limitations of existing 3C-based methods.
- Robert A. Beagrie
- , Antonio Scialdone
- & Ana Pombo
-
Letter |
 ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
The chromatin-reader protein ENL regulates oncogenic programs in acute myeloid leukaemia by binding via its YEATS domain to acetylated histones on the promoters of actively transcribed genes and recruiting the transcriptional machinery.
- Liling Wan
- , Hong Wen
- & Xiaobing Shi
-
Letter |
 Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping
Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping
The first genome-wide map of human chromatin conformation at the 1–3 nucleosome (50–500 base pair) scale, obtained using ionizing radiation-induced spatially correlated cleavage of DNA with sequencing (RICC-seq), which identifies spatially proximal DNA–DNA contacts.
- Viviana I. Risca
- , Sarah K. Denny
- & William J. Greenleaf
-
Letter |
 Structure and regulation of the chromatin remodeller ISWI
Structure and regulation of the chromatin remodeller ISWI
The crystal structures of ISWI, the catalytic subunit of several chromatin remodelling complexes, and its complex with a histone H4 peptide are reported.
- Lijuan Yan
- , Li Wang
- & Zhucheng Chen
-
Letter |
 Mechanism for DNA transposons to generate introns on genomic scales
Mechanism for DNA transposons to generate introns on genomic scales
The observations that introns are acquired in bursts and that exons are often nucleosome-sized can be explained by the generation of introns from DNA transposons, which insert between nucleosomes.
- Jason T. Huff
- , Daniel Zilberman
- & Scott W. Roy
-
Letter |
 Formation of new chromatin domains determines pathogenicity of genomic duplications
Formation of new chromatin domains determines pathogenicity of genomic duplications
Genomic duplications in the SOX9 region are associated with human disease phenotypes; a study using human cells and mouse models reveals that the duplications can cause the formation of new higher-order chromatin structures called topologically associated domains (TADs) thereby resulting in changes in gene expression.
- Martin Franke
- , Daniel M. Ibrahim
- & Stefan Mundlos
-
Letter |
 Allelic reprogramming of the histone modification H3K4me3 in early mammalian development
Allelic reprogramming of the histone modification H3K4me3 in early mammalian development
Three papers in this issue of Nature use highly sensitive ChIP–seq assays to describe the dynamic patterns of histone modifications during early mouse embryogenesis, showing that oocytes have a distinctive epigenome and providing insights into how the maternal gene expression program transitions to the zygotic program.
- Bingjie Zhang
- , Hui Zheng
- & Wei Xie
-
Letter |
 Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition
Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition
Three papers in this issue of Nature use highly sensitive ChIP–seq assays to describe the dynamic patterns of histone modifications during early mouse embryogenesis, showing that oocytes have a distinctive epigenome and providing insights into how the maternal gene expression program transitions to the zygotic program.
- John Arne Dahl
- , Inkyung Jung
- & Arne Klungland
-
Letter |
 Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos
Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos
Three papers in this issue of Nature use highly sensitive ChIP–seq assays to describe the dynamic patterns of histone modifications during early mouse embryogenesis, showing that oocytes have a distinctive epigenome and providing insights into how the maternal gene expression program transitions to the zygotic program.
- Xiaoyu Liu
- , Chenfei Wang
- & Shaorong Gao
-
Letter |
 Insights from biochemical reconstitution into the architecture of human kinetochores
Insights from biochemical reconstitution into the architecture of human kinetochores
Biochemical reconstitution of a synthetic human kinetochore with 21 protein subunits and centromeric nucleosomal DNA unveils fundamental principles of kinetochore organization and function.
- John R. Weir
- , Alex C. Faesen
- & Andrea Musacchio
-
Letter |
 The structural basis of modified nucleosome recognition by 53BP1
The structural basis of modified nucleosome recognition by 53BP1
A cryo-electron microscopy structure of the DNA damage repair protein 53BP1 bound to a nucleosome illuminates the way 53BP1 recognizes two types of histone modifications (a methyl group and a ubiquitin moiety), and provides insight into the highly specified recognition and recruitment of 53BP1 to modified chromatin.
- Marcus D. Wilson
- , Samir Benlekbir
- & Daniel Durocher
-
Letter |
 A core viral protein binds host nucleosomes to sequester immune danger signals
A core viral protein binds host nucleosomes to sequester immune danger signals
Here, a small core protein of human adenoviruses is shown to associate with histones, sequestering proteins on host chromatin and preventing inflammatory proteins from being released and triggering inflammation.
- Daphne C. Avgousti
- , Christin Herrmann
- & Matthew D. Weitzman
-
Letter |
 H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex
H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex
We have a limited understanding of how cells mark and identify newly replicated genomic loci that have a sister chromatid; here, unmethylated K20 in the tail of new histone H4 is shown to serve as a signature of post-replicative chromatin, which is specifically recognized by the homologous recombination complex TONSL–MMS22L.
- Giulia Saredi
- , Hongda Huang
- & Anja Groth
-
Article |
 The landscape of accessible chromatin in mammalian preimplantation embryos
The landscape of accessible chromatin in mammalian preimplantation embryos
An improved ATAC-seq approach is used to describe a genome-wide view of accessible chromatin and cis-regulatory elements in mouse preimplantation embryos, allowing construction of a regulatory network of early development that helps to identify key modulators of lineage specification.
- Jingyi Wu
- , Bo Huang
- & Wei Xie
-
Letter |
 AMPK–SKP2–CARM1 signalling cascade in transcriptional regulation of autophagy
AMPK–SKP2–CARM1 signalling cascade in transcriptional regulation of autophagy
An investigation into the nuclear events involved in autophagy regulation identifies the histone arginine methyltransferase CARM1 as a transcriptional co-activator of transcription factor TFEB; CARM1 levels are decreased by the SKP2-containing E3 ubiquitin ligase and increased during autophagy induction after nutrient starvation.
- Hi-Jai R. Shin
- , Hyunkyung Kim
- & Sung Hee Baek
-
Letter |
 Co-repressor CBFA2T2 regulates pluripotency and germline development
Co-repressor CBFA2T2 regulates pluripotency and germline development
A co-repressor protein, CBFA2T2, oligomerizes to stabilize its binding partner PRDM14 and the pluripotency factor OCT4 on chromatin, thus facilitating the transcriptional landscape underpinning the germline and pluripotent fate.
- Shengjiang Tu
- , Varun Narendra
- & Danny Reinberg
-
Article |
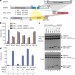 Structural basis for activity regulation of MLL family methyltransferases
Structural basis for activity regulation of MLL family methyltransferases
Crystal structures of the SET domains of MLL3 and a mutant MLL1 either unbound or complexed with domains from RBBP5 and ASH2L are determined; a combination of structural, biochemical and computational analyses reveals a two-step activation mechanism of MLL family proteins, which may be relevant for other histone methyltransferases.
- Yanjing Li
- , Jianming Han
- & Ming Lei
-
Letter |
 Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells
Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells
Genome-wide binding profiles for eight different chromatin remodellers in mouse embryonic stem (ES) cells are determined at single nucleosome resolution; each remodeller binds at specific nucleosome positions relative to the start of genes, and the same remodeller acts as a positive or negative regulator of transcription depending on the promoter chromatin organization and epigenetic marking of the gene it binds.
- Maud de Dieuleveult
- , Kuangyu Yen
- & Matthieu Gérard
-
Letter |
 Super-resolution imaging reveals distinct chromatin folding for different epigenetic states
Super-resolution imaging reveals distinct chromatin folding for different epigenetic states
Using super-resolution imaging to directly observe the three-dimensional organization of Drosophila chromatin at a scale spanning sizes from individual genes to entire gene regulatory domains, the authors find that transcriptionally active, inactive and Polycomb-repressed chromatin states each have a distinct spatial organisation.
- Alistair N. Boettiger
- , Bogdan Bintu
- & Xiaowei Zhuang
-
Letter |
 Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer
Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer
BET inhibitors that target bromodomain chromatin readers such as BRD4 are being explored as potential therapeutics in cancer; here triple-negative breast cancer cell lines are shown to respond to BET inhibitors and resistance seems to be associated with transcriptional changes rather than drug efflux and mutations, opening potential avenues to improve clinical responses to BET inhibitors.
- Shaokun Shu
- , Charles Y. Lin
- & Kornelia Polyak
-
Letter |
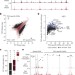 Competition between DNA methylation and transcription factors determines binding of NRF1
Competition between DNA methylation and transcription factors determines binding of NRF1
The relationship between DNA methylation and transcription factor binding was studied across the genome in mouse embryonic stem cells-the study reveals that the transcription factor NRF1 is methylation-sensitive and how physiological binding of NRF1 relies on local removal of DNA methylation.
- Silvia Domcke
- , Anaïs Flore Bardet
- & Dirk Schübeler
-
Letter |
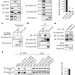 Autophagy mediates degradation of nuclear lamina
Autophagy mediates degradation of nuclear lamina
In response to cancer-associated stress, autophagy machinery mediates degradation of nuclear lamina components in mammals, suggesting that cells might degrade nuclear components to prevent tumorigenesis.
- Zhixun Dou
- , Caiyue Xu
- & Shelley L. Berger
-
Letter |
 Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage
Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage
At the initiation of DNA double-strand break repair, a number of ubiquitylation events occur; here, the RNF8 ubiquitin E3 ligase and the ubiquitin-conjugating E2 enzyme, UBC13, are shown to primarily modify H1-type linker histones, via a K63 linkage.
- Tina Thorslund
- , Anita Ripplinger
- & Niels Mailand
-
Letter |
 BET inhibitor resistance emerges from leukaemia stem cells
BET inhibitor resistance emerges from leukaemia stem cells
BET inhibitors that target bromodomain chromatin readers such as BRD4 are being explored as potential therapeutics in cancer; here, in a MLL–AF9 mouse leukaemia model, resistance to BET inhibitors is shown to emerge from leukaemia stem cells, and be partly due to increased Wnt/β-catenin signalling.
- Chun Yew Fong
- , Omer Gilan
- & Mark A. Dawson
-
Letter |
 Transcriptional plasticity promotes primary and acquired resistance to BET inhibition
Transcriptional plasticity promotes primary and acquired resistance to BET inhibition
BET bromodomain inhibitors are being explored as potential therapeutics in cancer; here, AML cells are shown to evade sensitivity to BET inhibition through rewiring the transcriptional regulation of BRD4 target genes such as MYC in a process that is facilitated by suppression of PRC2 and WNT signalling activation.
- Philipp Rathert
- , Mareike Roth
- & Johannes Zuber
-
Article |
 Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth
Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth
A ChIP-seq analysis of the DNA-binding properties of mutant gain-of-function p53 protein compared to wild-type p53 reveals the gain-of-function proteins bind to and activate a distinct set of genes including chromatin modifying enzymes such as the histone methyltransferase MLL; small molecular inhibitors of MLL function may represent a new target for cancers with mutant p53.
- Jiajun Zhu
- , Morgan A. Sammons
- & Shelley L. Berger
-
Letter |
 Single-cell chromatin accessibility reveals principles of regulatory variation
Single-cell chromatin accessibility reveals principles of regulatory variation
A single-cell method for probing genome-wide chromatin accessibility has been developed; the results provide insight into the relationship between cell-to-cell variation associated with specific trans-factors and cis-elements, as well insights into the relationship between chromatin accessibility and three-dimensional genome organization.
- Jason D. Buenrostro
- , Beijing Wu
- & William J. Greenleaf
-
Letter |
 Structural basis for retroviral integration into nucleosomes
Structural basis for retroviral integration into nucleosomes
Retroviruses such as HIV rely on the intasome, a tetramer of integrase protein bound to the viral DNA ends interacting with host chromatin, for integration into the host genome; the structure of the intasome as it interacts with a nucleosome is now solved, giving insight into the integration process.
- Daniel P. Maskell
- , Ludovic Renault
- & Peter Cherepanov
-
Letter |
 Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells
Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells
Transposable elements in mammalian genomes need to be silenced to avoid detrimental genome instability; here, the histone variant H3.3 is shown to have an important role in silencing endogenous retroviral elements.
- Simon J. Elsässer
- , Kyung-Min Noh
- & Laura A. Banaszynski
-
Letter |
 The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3
The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3
The mechanisms by which Xist, a long non-coding RNA, silences one X chromosome in female mammals are unknown; here a mass spectrometry-based approach is developed to identify several proteins that interact directly with Xist, including the transcriptional repressor SHARP that is required for transcriptional silencing through the histone deacetylase HDAC3.
- Colleen A. McHugh
- , Chun-Kan Chen
- & Mitchell Guttman
-
Article
| Open Access Transcription factor binding dynamics during human ES cell differentiation
Transcription factor binding dynamics during human ES cell differentiation
Lineage-specific transcription factors and signalling pathways cooperate with pluripotency regulators to control the transcriptional networks that drive cell specification and exit from an embryonic stem cell state; here, we report genome-wide binding data for 38 transcription factors combined with analysis of epigenomic and gene expression data during the differentiation of human embryonic stem cells into the three germ layers.
- Alexander M. Tsankov
- , Hongcang Gu
- & Alexander Meissner
-
Letter |
 Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation
Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation
Genome-wide localization and activity analysis of the de novo DNA methyltransferases DNMT3A and DNMT3B in mouse embryonic stem cells identifies overlapping and individual targeting preferences to the genome, including a role for DNMT3B in gene body methylation.
- Tuncay Baubec
- , Daniele F. Colombo
- & Dirk Schübeler
-
Letter |
 RNA helicase DDX21 coordinates transcription and ribosomal RNA processing
RNA helicase DDX21 coordinates transcription and ribosomal RNA processing
DEAD-box RNA helicase DDX21 is involved in both the transcription and RNA processing of ribosomal genes in human cells, sensing the transcriptional status of both RNA polymerase I and RNA polymerase II and associating with non-coding RNAs involved in ribonucleoprotein formation, possibly allowing for coordinated regulation of protein synthesis.
- Eliezer Calo
- , Ryan A. Flynn
- & Joanna Wysocka
-
Article |
 Synaptic, transcriptional and chromatin genes disrupted in autism
Synaptic, transcriptional and chromatin genes disrupted in autism
Whole-exome sequencing in a large autism study identifies over 100 autosomal genes that are likely to affect risk for the disorder; these genes, which show unusual evolutionary constraint against mutations, carry de novo loss-of-function mutations in over 5% of autistic subjects and many function in synaptic, transcriptional and chromatin-remodelling pathways.
- Silvia De Rubeis
- , Xin He
- & Joseph D. Buxbaum
-
Article |
 Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome
Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome
The crystal structure of the PRC1 ubiquitylation module bound to its nucleosome core substrate is determined, revealing how a histone-modifying enzyme achieves substrate specificity by recognizing nucleosome surfaces distinct from the site of catalysis, and uncovering a unique role for the ubiquitin E2 enzyme in substrate recognition.
- Robert K. McGinty
- , Ryan C. Henrici
- & Song Tan

 Nucleosome–Chd1 structure and implications for chromatin remodelling
Nucleosome–Chd1 structure and implications for chromatin remodelling