Featured
-
-
Article
| Open Access Single-cell DNA methylome and 3D multi-omic atlas of the adult mouse brain
Single-cell DNA methylome and 3D multi-omic atlas of the adult mouse brain
Methylome-based clustering and cross-modality integration with companion datasets from the BRAIN Initiative Cell Census Network enabled the construction of a 3D multi-omic genome atlas of the adult mouse brain featuring thousands of cell-type-specific profiles.
- Hanqing Liu
- , Qiurui Zeng
- & Joseph R. Ecker
-
Article |
 Establishment and function of chromatin organization at replication origins
Establishment and function of chromatin organization at replication origins
Genome-scale in vitro reconstitution of DNA replication through chromatin establishes a crucial role for the origin recognition complex in organizing nucleosome arrays that are crucial for the initiation of replication.
- Erika Chacin
- , Karl-Uwe Reusswig
- & Christoph F. Kurat
-
Article |
 Structural variants drive context-dependent oncogene activation in cancer
Structural variants drive context-dependent oncogene activation in cancer
Results are presented that indicate that alterations to gene regulatory three-dimensional architecture are a critical mechanism that enables structural variant-based oncogene activation in cancer genomes and sheds light on the essential elements for such gene activation events.
- Zhichao Xu
- , Dong-Sung Lee
- & Jesse R. Dixon
-
Article
| Open Access Cohesin-mediated loop anchors confine the locations of human replication origins
Cohesin-mediated loop anchors confine the locations of human replication origins
A study shows that the three-dimensional conformation of the human genome influences the positioning of DNA replication initiation zones, highlighting cohesin-mediated loop anchors as essential determinants of their precise location.
- Daniel J. Emerson
- , Peiyao A. Zhao
- & Jennifer E. Phillips-Cremins
-
Article |
 Phase separation drives aberrant chromatin looping and cancer development
Phase separation drives aberrant chromatin looping and cancer development
The NUP98–HOXA9 oncogenic fusion protein found in leukaemia undergoes phase separation in the nucleus, which helps to promote activation of leukaemic genes and to establish aberrant chromatin looping.
- Jeong Hyun Ahn
- , Eric S. Davis
- & Gang Greg Wang
-
Article |
 Defining genome architecture at base-pair resolution
Defining genome architecture at base-pair resolution
Micro Capture-C allows physical contacts to be determined at base-pair resolution, revealing that transcription factors have an important role in the maintenance of the contacts between enhancers and promoters.
- Peng Hua
- , Mohsin Badat
- & James O. J. Davies
-
Article |
 Loop extrusion as a mechanism for formation of DNA damage repair foci
Loop extrusion as a mechanism for formation of DNA damage repair foci
During the repair of double-stranded DNA breaks, cohesin mediates the extrusion of loops of DNA along which phosphorylated H2AX spreads to establish a repair zone.
- Coline Arnould
- , Vincent Rocher
- & Gaëlle Legube
-
Article |
 H1 histones control the epigenetic landscape by local chromatin compaction
H1 histones control the epigenetic landscape by local chromatin compaction
Experiments using a conditional triple-knockout mouse strain show that histone H1 regulates the activity of chromatin domains by controlling chromatin compaction, genome architecture and histone methylation.
- Michael A. Willcockson
- , Sean E. Healton
- & Arthur I. Skoultchi
-
Article |
 A map of cis-regulatory elements and 3D genome structures in zebrafish
A map of cis-regulatory elements and 3D genome structures in zebrafish
A comprehensive map of transcriptomes, cis-regulatory elements, heterochromatin structure, the methylome and 3D genome organization in the zebrafish (Danio rerio) enables identification of species-specific and evolutionarily conserved regulatory features, and provides a foundation for modelling studies on human disease and development.
- Hongbo Yang
- , Yu Luan
- & Feng Yue
-
Article
| Open Access Landscape of cohesin-mediated chromatin loops in the human genome
Landscape of cohesin-mediated chromatin loops in the human genome
A map of cohesin-mediated chromatin loops in 24 types of human cells identifies loops that show cell-type-specific variation, indicating that chromatin loops may help to specify cell-specific gene expression programs and functions.
- Fabian Grubert
- , Rohith Srivas
- & Michael Snyder
-
Article |
 Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion
Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion
Pax5 regulates contraction of the immunoglobulin heavy chain (Igh) locus—an essential step in V(D)J recombination—by promoting chromatin loop extrusion via repression of Wapl expression.
- Louisa Hill
- , Anja Ebert
- & Meinrad Busslinger
-
Article |
 Negative supercoil at gene boundaries modulates gene topology
Negative supercoil at gene boundaries modulates gene topology
The topoisomerase Top2 and the chromatin-binding protein Hmo1 maintain under-wound and over-wound DNA at different regions within a gene and thereby modulate the topology of genes.
- Yathish Jagadheesh Achar
- , Mohamood Adhil
- & Marco Foiani
-
Article |
 The structural basis for cohesin–CTCF-anchored loops
The structural basis for cohesin–CTCF-anchored loops
The crystal structure of the SA2–SCC1 subunits of human cohesin in complex with CTCF reveals the molecular basis of the cohesin–CTCF interaction that enables the dynamic regulation of chromatin folding.
- Yan Li
- , Judith H. I. Haarhuis
- & Daniel Panne
-
Article |
 Chromatin structure dynamics during the mitosis-to-G1 phase transition
Chromatin structure dynamics during the mitosis-to-G1 phase transition
Analysis of the dynamics of chromosome reorganization after exit from mitosis reveals the distinct but mutually influential forces that drive chromatin reconfiguration.
- Haoyue Zhang
- , Daniel J. Emerson
- & Gerd A. Blobel
-
Article |
 Stabilization of chromatin topology safeguards genome integrity
Stabilization of chromatin topology safeguards genome integrity
Super-resolution microscopy demonstrates how changes in the 3D organization of chromatin protect DNA against excessive degradation following damage.
- Fena Ochs
- , Gopal Karemore
- & Claudia Lukas
-
Letter |
 The fundamental role of chromatin loop extrusion in physiological V(D)J recombination
The fundamental role of chromatin loop extrusion in physiological V(D)J recombination
V(D)J recombination in B cells involves cohesin-mediated extrusion of chromatin loops to present DNA targets for cleavage and joining.
- Yu Zhang
- , Xuefei Zhang
- & Frederick W. Alt
-
Article |
 Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy
Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy
A disease model using cardiomyocytes derived from induced pluripotent stem cells of patients with mutated LMNA-related dilated cardiomyopathy reveals that the abnormal activation of the PDGF pathway is associated with the arrhythmic phenotypes of patients.
- Jaecheol Lee
- , Vittavat Termglinchan
- & Joseph C. Wu
-
Letter |
 Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors
Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors
Genetic deletion of mammalian chromatin remodelling complexes reveals that ISWI and SWI/SNF are required for binding of specific transcription factors and that ISWI regulates nucleosome positioning and nuclear organization in stem cells.
- Darko Barisic
- , Michael B. Stadler
- & Dirk Schübeler
-
Article |
 Visualizing DNA folding and RNA in embryos at single-cell resolution
Visualizing DNA folding and RNA in embryos at single-cell resolution
Optical reconstruction of chromatin architecture and multiplex RNA labelling traces the DNA path in single cells and its relationship to transcription.
- Leslie J. Mateo
- , Sedona E. Murphy
- & Alistair N. Boettiger
-
Letter |
 Multiplex chromatin interactions with single-molecule precision
Multiplex chromatin interactions with single-molecule precision
A strategy using droplet-based and barcode-linked sequencing captures multiplex chromatin interactions at single-molecule precision, and here provides topological insight into chromatin structures and transcription in Drosophila.
- Meizhen Zheng
- , Simon Zhongyuan Tian
- & Yijun Ruan
-
Article |
 LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice
LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice
Specific interchromosomal contacts in olfactory sensory neurons form a super-enhancer that controls the expression of a single olfactory receptor in each neuron.
- Kevin Monahan
- , Adan Horta
- & Stavros Lomvardas
-
Article |
 Two independent modes of chromatin organization revealed by cohesin removal
Two independent modes of chromatin organization revealed by cohesin removal
Depletion of chromosome-associated cohesin leads to loss of topologically associating domains in interphase chromosomes, without affecting segregation into compartments, and instead, it unmasks a finer compartment structure that reflects local chromatin and transcriptional activity.
- Wibke Schwarzer
- , Nezar Abdennur
- & Francois Spitz
-
Letter |
 Allelic reprogramming of 3D chromatin architecture during early mammalian development
Allelic reprogramming of 3D chromatin architecture during early mammalian development
A low-input Hi-C method is used to show that chromatin organization is markedly relaxed in pre-implantation mouse embryos after fertilization and that the subsequent maturation of 3D chromatin architecture is surprisingly slow.
- Zhenhai Du
- , Hui Zheng
- & Wei Xie
-
Letter |
 Phase separation drives heterochromatin domain formation
Phase separation drives heterochromatin domain formation
HP1a can nucleate into foci that display liquid properties during the early stages of heterochromatin domain formation in Drosophila embryos, suggesting that the repressive action of heterochromatin may be mediated in part by emergent properties of phase separation.
- Amy R. Strom
- , Alexander V. Emelyanov
- & Gary H. Karpen
-
Letter |
 Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition
Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition
Using a single-nucleus Hi-C protocol, the authors find that spatial organization of chromatin during oocyte-to-zygote transition differs between paternal and maternal nuclei within a single-cell zygote.
- Ilya M. Flyamer
- , Johanna Gassler
- & Kikuë Tachibana-Konwalski
-
Article |
 3D structures of individual mammalian genomes studied by single-cell Hi-C
3D structures of individual mammalian genomes studied by single-cell Hi-C
A chromosome conformation capture method in which single cells are first imaged and then processed enables intact genome folding to be studied at a scale of 100 kb, validated, and analysed to generate hypotheses about 3D genomic interactions and organisation.
- Tim J. Stevens
- , David Lando
- & Ernest D. Laue
-
Article |
 Complex multi-enhancer contacts captured by genome architecture mapping
Complex multi-enhancer contacts captured by genome architecture mapping
A technique called genome architecture mapping (GAM) involves sequencing DNA from a large number of thin nuclear cryosections to develop a map of genome organization without the limitations of existing 3C-based methods.
- Robert A. Beagrie
- , Antonio Scialdone
- & Ana Pombo
-
Letter |
 Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping
Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping
The first genome-wide map of human chromatin conformation at the 1–3 nucleosome (50–500 base pair) scale, obtained using ionizing radiation-induced spatially correlated cleavage of DNA with sequencing (RICC-seq), which identifies spatially proximal DNA–DNA contacts.
- Viviana I. Risca
- , Sarah K. Denny
- & William J. Greenleaf
-
Letter |
 Formation of new chromatin domains determines pathogenicity of genomic duplications
Formation of new chromatin domains determines pathogenicity of genomic duplications
Genomic duplications in the SOX9 region are associated with human disease phenotypes; a study using human cells and mouse models reveals that the duplications can cause the formation of new higher-order chromatin structures called topologically associated domains (TADs) thereby resulting in changes in gene expression.
- Martin Franke
- , Daniel M. Ibrahim
- & Stefan Mundlos
-
Article |
 The landscape of accessible chromatin in mammalian preimplantation embryos
The landscape of accessible chromatin in mammalian preimplantation embryos
An improved ATAC-seq approach is used to describe a genome-wide view of accessible chromatin and cis-regulatory elements in mouse preimplantation embryos, allowing construction of a regulatory network of early development that helps to identify key modulators of lineage specification.
- Jingyi Wu
- , Bo Huang
- & Wei Xie
-
Letter |
 Super-resolution imaging reveals distinct chromatin folding for different epigenetic states
Super-resolution imaging reveals distinct chromatin folding for different epigenetic states
Using super-resolution imaging to directly observe the three-dimensional organization of Drosophila chromatin at a scale spanning sizes from individual genes to entire gene regulatory domains, the authors find that transcriptionally active, inactive and Polycomb-repressed chromatin states each have a distinct spatial organisation.
- Alistair N. Boettiger
- , Bogdan Bintu
- & Xiaowei Zhuang
-
Letter |
 In situ structural analysis of the human nuclear pore complex
In situ structural analysis of the human nuclear pore complex
The most comprehensive architectural model to date of the nuclear pore complex reveals previously unknown local interactions, and a role for nucleoporin 358 in Y-complex oligomerization.
- Alexander von Appen
- , Jan Kosinski
- & Martin Beck
-
Letter |
 The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3
The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3
The mechanisms by which Xist, a long non-coding RNA, silences one X chromosome in female mammals are unknown; here a mass spectrometry-based approach is developed to identify several proteins that interact directly with Xist, including the transcriptional repressor SHARP that is required for transcriptional silencing through the histone deacetylase HDAC3.
- Colleen A. McHugh
- , Chun-Kan Chen
- & Mitchell Guttman
-
Article |
 Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome
Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome
The crystal structure of the PRC1 ubiquitylation module bound to its nucleosome core substrate is determined, revealing how a histone-modifying enzyme achieves substrate specificity by recognizing nucleosome surfaces distinct from the site of catalysis, and uncovering a unique role for the ubiquitin E2 enzyme in substrate recognition.
- Robert K. McGinty
- , Ryan C. Henrici
- & Song Tan
-
Letter |
 A high-resolution map of the three-dimensional chromatin interactome in human cells
A high-resolution map of the three-dimensional chromatin interactome in human cells
A novel approach to analyse high-depth Hi-C data provides a comprehensive chromatin interaction map at approximately 5–10 kb resolution in human fibroblasts; this reveals that TNF-α-responsive enhancers are already in contact with target promoters before signalling and that this chromatin looping is a strong predictor of gene induction.
- Fulai Jin
- , Yan Li
- & Bing Ren
-
Article
| Open Access The accessible chromatin landscape of the human genome
The accessible chromatin landscape of the human genome
An extensive map of human DNase I hypersensitive sites, markers of regulatory DNA, in 125 diverse cell and tissue types is described; integration of this information with other ENCODE-generated data sets identifies new relationships between chromatin accessibility, transcription, DNA methylation and regulatory factor occupancy patterns.
- Robert E. Thurman
- , Eric Rynes
- & John A. Stamatoyannopoulos
-
Article |
 A map of nucleosome positions in yeast at base-pair resolution
A map of nucleosome positions in yeast at base-pair resolution
A new technique for mapping nucleosomes genome-wide with single-base-pair accuracy, by chemical modification of engineered histones.
- Kristin Brogaard
- , Liqun Xi
- & Jonathan Widom
-
Article |
 BRCA1 tumour suppression occurs via heterochromatin-mediated silencing
BRCA1 tumour suppression occurs via heterochromatin-mediated silencing
- Quan Zhu
- , Gerald M. Pao
- & Inder M. Verma
-
-
Article |
 Mediator and cohesin connect gene expression and chromatin architecture
Mediator and cohesin connect gene expression and chromatin architecture
Gene activation may involve the formation of a DNA loop that connects enhancer-bound transcription factors with the transcription apparatus at the core promoter. But this process is not well understood. Here, two proteins, mediator and cohesin, are shown to connect the enhancers and core promoters of active genes in embryonic stem cells. These proteins seem to generate cell-type-specific DNA loops linked to the gene expression program of each cell.
- Michael H. Kagey
- , Jamie J. Newman
- & Richard A. Young
-
Letter |
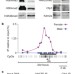 CpG islands influence chromatin structure via the CpG-binding protein Cfp1
CpG islands influence chromatin structure via the CpG-binding protein Cfp1
Most human gene promoters are embedded within CpG islands that lack DNA methylation and coincide with sites at which histone H3 lysine 4 is trimethylated (H3K4me3 sites). Here, a zinc-finger protein, Cfp1, is found to be associated with non-methylated CpG islands and H3K4me3 sites throughout the genome in the mouse brain. A primary function of non-methylated CpG islands might be to genetically determine the local chromatin modification state by interaction with Cfp1 and perhaps other CpG-binding proteins.
- John P. Thomson
- , Peter J. Skene
- & Adrian Bird
-

 Coordination of cohesin and DNA replication observed with purified proteins
Coordination of cohesin and DNA replication observed with purified proteins The Polycomb complex PRC2 and its mark in life
The Polycomb complex PRC2 and its mark in life Chromatin remodelling during development
Chromatin remodelling during development