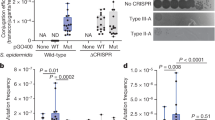
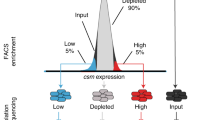
Abstract
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas systems have been adapted into a powerful genome-editing tool. The basis for the flexibility of the tool lies in the adaptive nature of CRISPR-Cas as a bacterial immune system. Here, we describe a protocol to experimentally demonstrate the adaptive nature of this bacterial immune system by challenging the model organism for the study of CRISPR adaptation, Streptococcus thermophilus, with phages in order to detect natural CRISPR immunization. A bacterial culture is challenged with lytic phages, the surviving cells are screened by PCR for expansion of their CRISPR array and the newly acquired specificities are mapped to the genome of the phage. Furthermore, we offer three variants of the assay to (i) promote adaptation by challenging the system using defective viruses, (ii) challenge the system using plasmids to generate plasmid-resistant strains and (iii) bias the system to obtain natural immunity against a specifically targeted DNA sequence. The core protocol and its variants serve as a means to explore CRISPR adaptation, discover new CRISPR-Cas systems and generate bacterial strains that are resistant to phages or refractory to undesired genes or plasmids. In addition, the core protocol has served in teaching laboratories at the undergraduate level, demonstrating both its robust nature and educational value. Carrying out the core protocol takes 4 h of hands-on time over 7 d. Unlike sequence-based methods for detecting natural CRISPR adaptation, this phage-challenge-based approach results in the isolation of CRISPR-immune bacteria for downstream characterization and use.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Deveau, H. et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 (2008).
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 (2011).
Garneau, J.E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Larson, M.H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196 (2013).
Makarova, K.S. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015).
Yosef, I., Goren, M.G. & Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 12, 5569–5576 (2012).
Wei, Y., Chesne, M.T., Terns, R.M. & Terns, M.P. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 43, 1749–1758 (2015).
Mojica, F.J.M., Díez-Villaseñor, C., García-Martínez, J. & Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 (2009).
Samson, J.E. & Moineau, S. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 4, 347–368 (2013).
Hynes, A.P., Villion, M. & Moineau, S. Adaptation in bacterial CRISPR-Cas immunity can be driven by defective phages. Nat. Commun. 5, 4399 (2014).
Bolotin, A., Quinquis, B., Sorokin, A. & Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005).
Levy, A. et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510 (2015).
Hynes, A.P., Labrie, S.J. & Moineau, S. Programming native CRISPR arrays for the generation of targeted immunity. MBio 7, e00202–e00216 (2016).
Díez-Villaseñor, C., Guzmán, N.M., Almendros, C., García-Martínez, J. & Mojica, F.J.M. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 10, 792–802 (2013).
Richter, C. et al. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 42, 8516–8526 (2014).
Datsenko, K.A. et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3, 945 (2012).
Shmakov, S. et al. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 42, 5907–5916 (2014).
Pride, D.T. et al. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res. 21, 126–136 (2011).
Lopez-Sanchez, M.J. et al. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 85, 1057–1071 (2012).
Labrie, S.J., Samson, J.E. & Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010).
Bondy-Denomy, J., Pawluk, A., Maxwell, K.L. & Davidson, A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013).
Martel, B. & Moineau, S. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 42, 9504–9513 (2014).
Magadán, A.H., Dupuis, M.-È., Villion, M. & Moineau, S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One 7, e40913 (2012).
van der Ploeg, J.R. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155, 1966–1976 (2009).
Cady, K.C., Bondy-Denomy, J., Heussler, G.E., Davidson, A.R. & O'Toole, G.A. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered Phages. J. Bacteriol. 194, 5728–5738 (2012).
Vorontsova, D. et al. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res. 43, 10848–10860 (2015).
Swarts, D.C., Mosterd, C., van Passel, M.W.J. & Brouns, S.J.J. CRISPR interference directs strand specific spacer acquisition. PLoS One 7, e35888 (2012).
Erdmann, S. & Garrett, R.A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 85, 1044–1056 (2012).
Li, M., Wang, R., Zhao, D. & Xiang, H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492 (2014).
Hooton, S.P.T. & Connerton, I.F. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front. Microbiol. 5, 744 (2014).
Erdmann, S., Le Moine Bauer, S. & Garrett, R.A. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 91, 900–917 (2014).
Heler, R. et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202 (2015).
Rao, C. et al. Active and adaptive Legionella CRISPR-Cas reveals a recurrent challenge to the pathogen. Cell. Microbiol. 18, 1319–38 (2016).
Silas, S. et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase–Cas1 fusion protein. Science 351, 929–932 (2016).
Acknowledgements
We recognize members of our team who were involved in the initial development of these protocols, namely M.-È. Dupuis, J. Garneau, S. Labrie, A. Magadán, B. Martel and M. Villion. M. Sabri was involved in the initial implementation of the protocol at the undergraduate level. We thank A. Renaud for assistance with materials to generate figures.
A.P.H. is supported by a scholarship from the National Science and Engineering Research Council of Canada (NSERC). M.-L.L. is supported by scholarships from the Fonds de Recherche du Québec—Nature et Technologies (FRQNT), Novalait and Op+Lait. S.M. acknowledges funding from the Natural Sciences and Engineering Research Council of Canada (Discovery program), Canadian Institutes of Health Research (Team Grant–Emerging: Novel Alternatives to Antibiotics) and Danisco/DuPont. S.M. holds a T1 Canada Research Chair in Bacteriophages.
Author information
Authors and Affiliations
Contributions
A.P.H. developed Variants 1 and 3. M.-L.L. was in the first undergraduate cohort to perform the protocols, and aided in their implementation in the following 2 years. L.T. and M.F. implemented the core protocol in undergraduate laboratories in all 3 years. H.D. helped develop the initial core protocol, and implemented it in undergraduate laboratories for 2 years. S.M. and D.M.T. were involved in the development of the core protocol and the three variants, and implemented the core protocol in undergraduate laboratories in the first year. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hynes, A., Lemay, ML., Trudel, L. et al. Detecting natural adaptation of the Streptococcus thermophilus CRISPR-Cas systems in research and classroom settings. Nat Protoc 12, 547–565 (2017). https://doi.org/10.1038/nprot.2016.186
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.186
This article is cited by
-
A truncated anti-CRISPR protein prevents spacer acquisition but not interference
Nature Communications (2022)
-
Competition and coevolution drive the evolution and the diversification of CRISPR immunity
Nature Ecology & Evolution (2022)
-
A mutation in the methionine aminopeptidase gene provides phage resistance in Streptococcus thermophilus
Scientific Reports (2019)
-
Recombination between phages and CRISPR−cas loci facilitates horizontal gene transfer in staphylococci
Nature Microbiology (2019)
-
Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.