Key Points
-
Provides an overview on basic composition and distribution of oral microbiota.
-
Elucidates the underlying mechanisms of endogenous and exogenous factors on oral microbiota and oral health.
-
Reviews oral microbiota and its implications for systemic diseases.
-
Summarises the improvement of clinical diagnosis and treatment based on microbial community information.
Abstract
Human oral microbiota is the ecological community of commensal, symbiotic, and pathogenic microorganisms found in the oral cavity. Oral microbiota generally exists in the form of a biofilm and plays a crucial role in maintaining oral homeostasis, protecting the oral cavity and preventing disease development. Human oral microbiota has recently become a new focus research for promoting the progress of disease diagnosis, assisting disease treatment, and developing personalised medicines. In this review, the scientific evidence supporting the association that endogenous and exogenous factors (diet, smoking, drinking, socioeconomic status, antibiotics use and pregnancy) modulate oral microbiota. It provides insights into the mechanistic role in which oral microbiota may influence systemic diseases, and summarises the challenges of clinical diagnosis and treatment based on the microbial community information. It provides information for noninvasive diagnosis and helps develop a new paradigm of personalised medicine. All these benefit human health in the post-metagenomics era.
Similar content being viewed by others
Introduction
The oral cavity is a connection channel between outside environments and the respiratory tract and digestive tract. It provides an appropriate temperature, humidity, and nutrition for microorganism colonisation. The human oral microbiome has been extensively studied as part of the Human Microbiome Project. The oral microbiome has an essential role in maintaining a normal oral ecological balance and in the development of oral diseases. There is abundant evidence supporting the theory that endogenous and exogenous factors are closely related to oral microbiota and systemic diseases.1,2 Studies on dietary behaviours demonstrate a fundamental aspect of the oral disease paradigm.3 Lifestyles and diets including smoking, alcohol drinking and consuming spicy food, and antibiotic treatments can persistently alter commensal microbial communities.4 The resultant microbial disturbances may increase pathogen susceptibility.5
The disturbance of the oral microbiota–ecology balance in the host usually causes a series of oral infectious diseases including dental caries, apical periodontitis, periodontal diseases, pericoronitis, and craniofacial bone osteomyelitis. Oral microbiota is also associated with several systemic diseases, namely cardiovascular disease, pneumonia, heart disease, rheumatoid arthritis, pancreatic cancer, colorectal cancer, oesophageal cancer, stroke, and adverse pregnancy outcomes. Accordingly, oral microbiota has been considered as a potential biomarker for human diseases. Relationships between oral microbiota and systemic diseases are essential and need to be elucidated, in order to provide a reasonable diagnosis basis for disease prevention and treatments.
This article mainly discusses the mechanisms for how endogenous and exogenous factors modulate oral microbiota, provides insights into their roles in the influence of oral microbiota on systemic diseases, and summarises the challenges for clinical diagnosis and treatment.
Basic composition and distribution of oral microbiota
The oral microbiome can be classified into core microbiome and variable microbiome. The core microbiome is similar for all individuals and comprised of the predominant species at different sites of the healthy body. The variable microbiome is different between individuals in response to unique lifestyles and phenotypic and genotypic determinants.
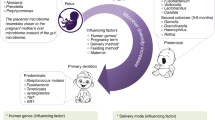
For newborns, within five minutes of birth, bacterial communities in the oral cavity and other body habitats are very similar to each other.6 Types of microorganisms are closely decided by the delivery mode.7 In addition, the mother's oral microbiota is the most important source of infants' and young children's oral microbiota by successful vertical transmission.7,8 As ageing continues, babies and children form a wide variety of oral microorganisms in response to different diets, lifestyles, environments and so on.9
The oral cavity contains over 700 microbial species as well as commensal and opportunistic bacteria, archaea, fungi, protozoa, and viruses.10,11 Every species plays its particular role and strongly interacts with the other species and the host.7 Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria are probably most significant for oral health.12 The major genera with the largest representation in oral cavities include the following: Streptococcus, Prevotella, Haemophilus, Rothia, Veillonellaceae, Neisseria, Fusobacterium and Porphyrin.13 Recently, primer pairs have been developed to make phylum-selective 16S rRNA clone libraries. In the libraries, species (Chloroflexi, Synergistetes, Chlorobi, Gracilibacteria, Saccharibacteria, and others) are identified from the lesser known oral phyla or candidate divisions.14,15
The salivary microbial groups are stable for a short term, although there are significant differences in the oral microbial groups affected by a variety of factors.16,17 There are 11 human microbial habitats in the oral cavity, including hard palate, tongue dorsum, saliva, palatine tonsils, throat, buccal mucosa, keratinised gingiva, supra-gingival plaque, subgingival plaque, dentures, and lips. The 11 habitats have been shown to contain different core microorganisms when sampled from more than 200 healthy people with high throughput sequencing methods as listed in Table 1.7,13,18,19 Microbiomes from the same location on the body are more similar among different individuals than those from different locations on the same individual.12,20 Within these oral habitats, 13 (tongue dorsum) to 19 (hard palate) bacteria phyla were described, including 185 (tongue dorsum) to 322 (throat) genera.7 After many years of research, there are still new discoveries due to the microbial diversity.
Endogenous and exogenous factors affecting oral microbiota
In healthy individuals, oral microbiome balance is regarded as dynamic because it changes in response to endogenous and exogenous factors. Human lifestyle and experiences can quickly and profoundly change the stability of microbial communities associated with the host.5 In other words, host lifestyle, physiology, genotype, pathobiology, environment, immune system, transient community members, and socioeconomic status are generally considered as important factors in the multifactorial background of oral diseases and systemic diseases.1,21,22,23,24 However, the underlying mechanisms of these factors on oral microbiota and oral health are not yet fully elucidated.
Diets
The change of dietary macronutrients and diet type can lead to a shift of the oral microbiome and diseases. Nutrients, such as sugars, fats and vitamins, play important roles in the oral microbiome. In severe early childhood caries, sugar-rich diets and frequent snacks show the highest associations with Streptococcus mutans (S. mutans) and Fusobacterium nucleatum (F. nucleatum).25,26 Saturated fatty acids (SFA) and vitamin C intakes are consistently correlated with alpha (within-subjects) diversity indexes in both richness and diversity.27 The higher the SFA intake, the higher the relative abundance of fusobacteria (Leptotrichiaceae) and betaproteobacteria. Vitamin C and the other intake-related vitamins, for example, B vitamins and vitamin E, exhibited positive correlations with the population of fusobacteria.27 Adler and his co-workers also found that cats on dry-food diets showed very high diversity in oral microbiome, especially with a higher abundance of Porphyromonas spp.28
Dietary modification with increased fibrous foods and dairy products and decreased fatty and sugary foods has been advised to maintain a normal oral ecological balance.29,30,31
Smoking
Cigarettes are rich in bacterial diversity, harbouring a variety of microorganisms from environmental bacteria and commensals to potential oral pathogens. Bacteria presenting in the cigarette could be transferred to the mouths of smokers even before the cigarette is lit.32 Some of these bacteria including Bacillus spp. and Clostridium spp. could survive the burning/smoking process, be inhaled by smokers and other exposed individuals, and colonise the oral cavity.33
Additionally, several other potential mechanisms also reveal how smoking alters oral microbial ecology, including increasing the acidity of saliva, depleting oxygen, antibiotic effects, influencing oral bacterial adherence to mucosal surfaces, and impairing host immunity.34,35 For example, the oxygen deprivation hypothesis proves that smoking creates an environment favouring strict or facultative anaerobes over strict aerobes.36 The genus Streptococcus, Veillonella and Actinomyces are facultative or obligate anaerobes. Conversely, aerobes such as Neisseria subflava and Corynebacterium are depleted in smokers,1,36 consistent with previous studies.37,38
Drinking
The influence of red liquor and wine on the oral microbiota is different. Liquor could lead to an increase in the concentration and number of gram-positive bacteria, such as S. mutans.39 Oral bacteria converts ethanol to acetaldehyde, which is a toxin and recognised human carcinogen.40 The production of acetaldehyde might also directly result in inhibition of fusobacteria.41 Specific impurities, contaminants, N-nitrosodiethylamine, and polycyclic aromatic hydrocarbons generated in the fermentation, distillation or maturation processes also change the oral environment and affect certain species.42 However, the regular and moderate consumption of red wine does not change the overall diversity and stability of representative bacterial groups of the human saliva.43 Furthermore, the synthetic mixtures of the organic acids (succinic, malic, lactic, tartaric, citric, and acetic acid) in red and white wines are active against oral streptococci responsible for caries development and Streptococcus pyogenes responsible for pharyngitis.44 These suggest that moderate drinking of red and white wines can enhance oral health.
Antibiotics use
Antibiotics are a mainstay of treatment for bacterial infections worldwide. Antibiotics influence bacterial growth curves and this is why they are used to kill pathogens. Bactericidal antibiotics directly kill the bacteria, while bacteriostatic antibiotics inhibit their growth. Many studies report that antibiotics such as azithromycin, amoxicillin clindamycin and ciprofloxacin affect the amount and diversity of oral microbes.45,46 Some general changes can be observed such as an immediate decrease in actinobacteria count in throat.31 The reported data also demonstrate that the oral microbiome functions (microbe metabolic activity, microbial gene expression and protein synthesis) were also drastically changed as a direct consequence of antibiotic treatments. For example, antibiotics might damage and/or destruct the bacterial cells and consequently decrease their enzymatic activity.47 The extent to which our oral microbiota changes after an antibiotic intervention depends not only on the chemical nature of the antibiotic used to treat specific infections, but also on the type of administration, duration and dose, as well as the level of resistance that each microbiota develops.48 Therefore, the establishment of new drug-based therapeutic strategies would require multi-variable analysis.
Socioeconomic status
Among the factors affecting oral health, socioeconomic status (SES) is the important factor that should not be neglected. At present, little is known about the influence of differences in SES on the composition of the oral microbiome. Studies report that these differences are reflected by the bacterial profiles of saliva. Megasphaera micronuciformis, Veillonella atypical, Veillonella parvula, Rothia mucilaginosa, Prevotella histicola, Fusobacterium periodontium, Granulicatella adiacens and Tannerella forsythia were abundant in the high socioeconomic status group, while Aggregatibacter segnis, Achromobacter xylosoxidans and Neisseria cluster II were abundant in the low socioeconomic group.21
The SES of each family determines the choices of family members' educational attainment, health concepts, hygiene habits, dietary patterns and medical services. Therefore, Chu et al. believed that the oral microbiome might vary considerably over time because of SES and the phenomenon of reducing oral microbial diversity was disproportionately prevalent in low-SES neighbourhoods.49
Pregnancy
Variations in oral microbiota in pregnancy have been observed. In the early stages of pregnancy, the total number of cultivated microbes in pregnant women increase significantly. For example, the abundance of P. gingivalis and Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans) in the gingival sulcus were significantly higher than that in the non-pregnant group, whereas Prevotella intermedia (P. intermedia) and F. nucleatum did not change.50 From weeks 12 to 28 of pregnancy, no changes occur. During late pregnancy, Candida species were more frequently detected.
Total bacterial counts decreased postpartum. The most dramatic microbial changes were the decrease of species including Capnocytophaga ochracea, Capnocytophaga sputigena, Eubacterium saburreum, Fusobacterium nucleatum naviforme, Fusobacterium nucleatum polymorphum, Leptotrichia buccalis, Parvimonas micra, P. intermedia, Prevotella melaninogenica, Staphylococcus aureus (S. aureus), Streptococcus anginosus, Streptococcus intermedius, S. mutans, Streptococcus oralis, Streptococcus sanguinis, Selenomonas noxia, and Veillonella parvula, while the abundance of Neisseria mucosa was found to increase significantly over time.51 Changes in pregnancy (especially physiological condition and female hormones) have a significant impact on the oral microbiota, and may promote the colonisation of various microorganisms, especially periodontal pathogens, that may be a risk factor for the health of pregnant women.52
Oral bacteria and systemic diseases
Increasingly, evidence suggests that specific bacterial infections promote development of certain diseases. Accordingly, this section mainly summarises the relationships between oral bacteria and systemic diseases. It also provides a deep insight into the mechanistic role in the influence of oral microbiota on cancers and inflammatory diseases.
Oral bacteria and cancers
Oesophageal cancer
Oesophageal cancer is the eighth most frequent tumour and sixth leading cause of cancer death worldwide.55 The latest study showed that oral bacteria might increase the risk of oesophageal cancer. Immunohistochemically, Porphyromonas gingivalis (P. gingivalis) has been detected in 61% of cancerous tissues, 12% of adjacent tissues, and 0% of normal oesophageal mucosa. In addition, lysine-specific gingipain distribution and P. gingivalis 16S rDNA were also researched. These findings were pioneering in proving that P. gingivalis infected the oesophageal epithelium of oesophageal cancer patients. The infection was observed in association with the progression of oesophageal cancer, and could be an important biomarker for this disease. Furthermore, eradication of a common oral pathogen might help to reduce the burden of oesophageal cancer.56
Colorectal cancer
Fusobacteria, which are from the mouth, cause excessive immune responses and turn on cancer growth genes. The microbes have been linked with colorectal cancer.57 Fusobacteria gather massively in adenomas – a benign bowel growth that will become cancerous as time goes on. The polymicrobial nature of oral biofilms and the asaccharolytic metabolism of many of these species helps them live well in the microenvironment of colonic lesions.58 By attracting special immune cells, fusobacteria invade the bowel and set off an inflammatory response that could accelerate the formation of colorectal tumours. Fusobacteria have specific surface molecules assisting them to attach and invade human colorectal cancer cells. In colorectal cancer, F. nucleatum has been demonstrated to expand myeloid-derived immune cells, strongly inhibit T-cell proliferation or activation, and induce T-cell apoptosis.59 Periodontal diseases including tooth loss might increase systemic inflammation, lead to immune dysregulation, and alter gut microbiota, and therefore possibly influence colorectal carcinogenesis.60 However, oral bacteria F. nucleatum could protect all sorts of tumour cells from being killed by immune cells.61 The Fap2 protein of F. nucleatum directly interacts with TIGIT(T-cell immunoglobulin and ITIM domain), leading to the inhibition on NK cell cytotoxicity and T-cell activities.61 This was a tumour-based immune evasion mechanism that was bacteria-dependent, wherein F. nucleatum bound tumours were protected from NK-mediated killing and immune cell attack due to an interaction between the Fusobacterial protein Fap2 with the immune cells inhibitory receptor TIGIT. F. nucleatum adhered to various tumour cells. NK cells clustered around F. nucleatum coated tumour cells. F. nucleatum exerted its inhibitory effect on TIGIT through the immunoreceptor tail tyrosine (ITT)-like and the immunodominant tyrosine-based inhibitory (ITIM) motifs located in TIGIT cytoplasmic tail. TIGIT was expressed in tumour infiltrating lymphocytes (TILs) found within colon adenocarcinoma and F. nucleatum inhibited the activity of these TILs in a Fap2-dependent manner. Furthermore, the Fap2 protein of F. nucleatum could inhibit the activity of T-cells presenting in the peripheral blood, such as interferon-γ secretion. These discoveries can lead to a better early diagnosis technique and new strategy for the treatment of cancer patients. Furthermore, the correlation of Fusobacterium with T-cells and microRNA expressions still needs to be clarified in colorectal cancer.
Pancreatic cancer
Pancreatic cancer is the fourth leading cause of cancer-related death and a serious threat to human health. Oral pathogens, especially P. gingivalis and Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans) were associated with a high risk of pancreatic cancer.62 Microbial cells released from the biofilm through the epithelium and spread systemically via the blood circulation. Some of the bacteria isolated from pancreatic tissues were members of the oral microbiome.63 P. gingivalis, one of the aetiological factors in pancreatic cancer, had the ability to escape host response and impair innate immunity, subsequently strengthening the favourable inherent environment for bacterial overgrowth, which in turn might mediate the microbial community and promote the conversion from a symbiotic state to a dysbiotic state. All above changes might cause high levels of inflammation in pancreatic cancer.64 Nitrosamines in the oral cavity that raised levels of major oral bacteria (P. gingivalis), could induce and promote a rapid development of pancreatic cancer. Therefore, the oral bacteria (P. gingivalis and A. actinomycetemcomitans) can be a good candidate as an effective biosensor for early diagnosis of pancreatic cancer.
Oral bacteria and inflammatory diseases
Atherosclerosis
Accurate and early diagnosis of cardiovascular diseases will greatly improve the survival rate of patients. Oral microbiota such as S. mutans, P. gingivalis, and Gemella haemolysans (G. haemolysans) may play a role in cardiovascular disease.65,66,67
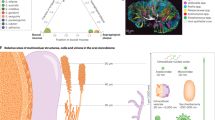
S. mutans could contribute directly to atherosclerosis by disrupting endothelial cell function, one of the earliest indicators of cardiovascular diseases.65 S. mutans is a major pathogen for dental caries. Oral S. mutans induced intracerebral haemorrhage experimentally and affected cerebral microbleeds.65 A significant correlation of cnm-positive S. mutans was observed with hypertensive intracerebral haemorrhage and deep cerebral microbleeds.68 G. haemolysans was simultaneously found in atherosclerotic and oral plaques of the elderly without periodontitis.66 As shown in Figure 1, there are three theories about bacteriology, inflammation, and immunology to explain the relationships between periodontal diseases and cardiovascular diseases.
In the early stage of infection, epithelial cells responded strongly to P. gingivalis by producing IL-6, INF-γ, or TNF-α, causing local tissue destruction. Subsequently, bacteria and virulence factors (for example, gingipains, lipopolysaccharide or fimbriae) entered into the bloodstream through degraded gingival tissues and activated endothelial cells, and produced inflammatory reaction under the action of pro-inflammatory mediators (for example, IL-8, MCP-1), growth factors, differentiation factors, cell-adhesion molecules and toll-like receptors. Eventually, P. gingivalis stimulation could shift endothelial cells toward a pro-thrombotic state (Fig. 1a).69
Furthermore, virulence factors accelerate the development of atheromatous plaque. Infection of epithelial cells by periodontal bacteria stimulated the production of proinflammatory cytokines (TNF-α, IL-1β, IL-6, and PGE2). These cytokines entered the blood circulation and affected cells in atheromatous plaques, leading to the development of atherosclerosis (Fig. 1c).67 Especially during developing periodontal diseases, both these monocytic hyper-inflammatory phenotypes amplify the inflammatory process and cross-reactivity inducing destruction of host cells promotes the development of atheromatous plaques (Fig. 1b).70,71
Pneumonia
Aspiration of bacteria from the oral cavity into the lower airway was possible since the surfaces of oral cavity were contiguous with those of the trachea and lower airway.72 Oral bacteria continuously flowed into the lungs, and the lungs exhausted the bacteria through ciliary actions and coughing.73 The lungs are constantly exposed to diverse communities of microbes from the oropharynx, and novel culture-independent techniques of microbial identification have revealed that the lungs, previously considered sterile in health, harbor diverse communities of microbes.
Streptococcus, Prevotella and Veillonella were the most common bacteria in healthy lungs. The microbial density in lungs was less than 1/1000 of that in the oral cavity, mainly because the lungs had no mucosa suitable for forming bacterial ecology.74,75 Control of oral biofilm formation could reduce the numbers of potential respiratory pathogens in the oral secretions, which in turn could reduce the risk for pneumonia.72 Recently, the concept of lung specific microbial groups had been accepted, but oral bacteria were still derived as a risk factor for ventilator-associated pneumonia. Under poor oral hygiene conditions, pathogens were easy to colonise in the oral cavity including methicillin resistant S. aureus, Pseudomonas aeruginosa, and ten genera of gram-negative bacilli. Subsequent aspiration would deposit these bacteria (especially anaerobic organisms derived from the gingival crevice) and inflammatory products (associated with periodontal disease) into the lower airway, thereby increasing the risk of lung infection.76,77
Heart disease
To date, the accumulated epidemiological evidence supported an association between oral bacterial diseases (such as periodontal diseases) and coronary artery disease (CAD). The present studies confirmed that five oral commensal bacteria (Campylobacter rectus, P. gingivalis, Porphyromonas endodontalis, P. intermedia, Prevotella nigrescens) were unique to coronary artery disease when compared with several non-cardiac disorders.78 And the presence of A. actinomycetemcomitans in the subgingival area was associated with an almost two fold risk of angiographically confirmed stable CAD.79 This suggested a special role for A. actinomycetemcomitans in CAD, other than only as a pathogen associated with periodontitis. Studies on the infection mechanisms between oral bacteria and CAD were essential for providing some clues for medicinal treatments in clinic. As depicted in Figure 2, many oral microbes that secreted proteins, peptides and proteases lived in the gingival crevice. These secretory peptides and proteases were likely responsible for altering the host actin cytoskeleton in the gingival epithelium leading to oral microbial entry into the bloodstream system. Upon gaining entry into the coronary vasculature, these migratory bacteria could form biofilm structures within atherosclerotic plaques and caused CAD. These secreted proteins could also activate the immune system causing inflammation. For example, cytokine-mediated (IL-6 and IL-8) inflammation was associated with CAD. Furthermore, certain proteases caused an inflammatory response by activating the complement system.
Rheumatoid arthritis
Rheumatoid arthritis (RA) is a systemic, inflammatory autoimmune disease. Most clinical studies involving specific oral microorganisms as triggers for RA were only dependent on serological detection methods. Rheumatoid factors had been identified as autoantibodies that reacted to the IgG molecule in the Fc region, and these antibodies could be the IgM, A, G or E epitopes. P. gingivalis proteinase was responsible for the epitope development in the RF Fc region. A previous study identified the lysine and arginine amino acid sequences for the Fc region of the IgG molecule; because P. gingivalis specifically decomposed lysine and arginine, the IgG3 CH2 and CH3 domains processed by P. gingivalis proteinase became powerful targets for the RF produced by rheumatoid cells.80 It was also found that the microbiome of patients with RA was similar to that of healthy subjects with similar periodontal status with the multiplexed-454-16S rRNA pyrosequencing method; however, specific Prevotella and Leptotrichia were only found in patients with new-onset RA, and anaeroglobus geminatus was correlated with the presence of peptidyl-arginine deiminase and rheumatoid factors, and with periodontitis.23 Another large-scale study using metagenomic shotgun sequencing identified compositional and functional alterations in RA-associated oral microbiomes, which were partly resolved by disease-modifying antirheumatic drugs treatments.81 Thus, all these data approaches suggested that microbiome composition could be important in the prognosis and diagnosis of RA.
The improvement of clinical diagnosis and treatment based on microbial community information
Databases of bacterial compositions generally identified by using high-throughput sequencing methods of the microbiome will facilitate advanced functional studies on genomics, transcriptomics, and the metabolomics of both host and pathogens. Such analysis can provide deep insights into the activity of the microbes, the relationship of the host and microbes, and potential causative mechanisms. The challenges of clinical diagnosis and treatment based on the microbial community information are still waiting to be conquered.
The standardisation of clinical samples
Standardisation of sampling plans implies that (a) the design elements of the sampling plan must be considered in any standardisation process and (b) the elements are selected to maximise performance.82 Oral micro-ecosystem is a complex system. Its microbial community species composition and genetic types are significantly different to the ecological sites and are even within the same site. These differences are further governed by a variety of host factors, including gene, health state, age, gender, dentition status, life-style, socioeconomic status, mobile phone use, living area, and religion.
Previous studies using high-throughput analysis techniques had observed that the oral cavity was a highly heterogeneous ecological system containing significantly different microbial communities. The Firmicutes was the dominant bacterium of salivary and dental mucosa, while Proteobacteria, Firmicutes, Bacteroidetes and Fusobacteria were the dominant bacteria of the dental plaque. More importantly, the oral microbial structure varied with age and dentition status.83 The results indicated that sampling process, sampling parts and the age of the sample objects were crucial to collect the accurate, systematic, and reproducible results.
Therefore, developing a uniform sampling plan to be used by all researchers is extremely important. The factors mentioned above should be taken into account in the standardisation of oral clinical samples so the errors can be effectively reduced. Furthermore, the standardisation of clinical samples also should be that:
-
The oral site needs to be delimited before sampling
-
Objective sampling should represent the entire oral ecosystem or site
-
Sampling should consist of the same small subsamples
-
Sampling using large-size samples implies that the selection of the sample site is representative
-
The sampling unit should be large enough for efficient statistical processing.
Analysis and processing of big data in an oral microbial community
Microbial big data are generated by high throughput sequencing, for functional prediction, biological classification of species, and gene analysis. They have rapidly developed into a hot topic that attracts extensive attention from academia, industry, and governments around the world. Although enhanced by the contents of the Human Oral Microbiome Database, the explosive growth of data presents us with grand challenges (namely, data complexity, computational complexity, and system complexity). The single data analysis process (Table 2) has some limitations, and does not meet the need for deep mining of microbial big data.
The lack of corresponding bioinformatics tools for reducing sequencing cost, optimising the analysis process, increasing specificity and sensitivity of biological community information and analysis method of sorting and digging large medical data are still the major bottleneck in the era of big data.
The following work may improve this situation:
-
Developing a set of interoperable data analysis tools that can run on different computing platforms. This will effectively improve the reliability and comparability of data analysis
-
Combining the data of electronic health records and genome data can help effectively explore the pathogenesis and therapeutic effect of diseases
-
Functional studies including genomics, transcriptomics and metabolomics of both host and oral pathogens.
This specific microbial application and analysis is in an exciting phase of research. Such analysis could guide researchers to develop new therapies that target key mechanisms. These are very crucial in advancing the personalised diseases early warning service for personalised diseases based on the oral microbiome.
Further verification of the cross-sectional study
More and more data show that the oral microbiome is related to dental diseases, cardiovascular disease and others. In these cross-sectional studies, various factors such as individual gene and bacterial variation influence the cross-sectional data and reduce reliability and accuracy of flora mapping. More importantly, the pure cross-sectional studies only provide the correlation of microbial community and diseases rather than clear their 'causality'. Host and microbiota have significant heterogeneity in various stages of the disease development. If a large number of data of genome, transcriptome, proteome, and metabolome are associated with clinical data such as clinical manifestations, pathology, biochemical markers and immune indicators, it looks forward to making clear the 'causality' of the core microbial group and disease, and crediting an oral microbiota-based prediction model to develop a new paradigm of personalised medicine.
Trends of treatment without antibiotics
Due to antibiotic overuse, the emergence of drug-resistant strains and frequent recurrence of the disease in affected individuals are increasing challenges in antifungal therapy. Moreover, indiscriminate use of antibiotics affects the delicate balance between normal flora and host. Beneficial bacteria are also eliminated, depriving the host from their beneficial effects. This has prompted the need for an alternative therapeutic and prevention strategy. Antibodies, vaccine, antimicrobial peptides, probiotics, prebiotics, synbiotics, and arginine become alternative therapeutic options, as illustrated in Table 3.
Antimicrobial peptides, probiotics combined with prebiotics and the screening probiotics, and arginine may assist or replace antibiotic treatments for oral microbial problems and in turn prevent systemic diseases. In the near future, a rapidly increasing body of knowledge promises to indicate more targeted applications of probiotics. It still needs to clearly determine which organisms are beneficial and play a preventive or therapeutic role. For those that can duly be termed probiotics, a variety of applications have to be defined more precisely than before.
Summary
Oral microbiota is an important intermediate link, causing different oral and overall health in the body under the influence of changes in a variety of factors. Once the microbiota balance has been disturbed, it may result in oral and even systemic diseases. Although a number of causes including infectious pathogens or use of antibiotics can lead to a disruption of microbial equilibrium, the role of our diet, nutrition, lifestyle and socioeconomic status is crucial.
In addition, observation of oral microbiota is a major indicator for the occurrence, development, and prognosis of disease. It has been verified that the microbiome is related to human physiology and pathology. An oral microbiota-based prediction model can provide the basis for noninvasive diagnosis and facilitate the development of a new paradigm of personalised medicine. All these benefit human health in the post-metagenomics era.
Funding sources
This work was supported by The National Science Foundation [grant numbers 31371809, 31771956], the “Shu Guang” project of Shanghai Municipal Education Commission and Shanghai Education Development Foundation [grant number 15SG42] and Natural Science Foundation of Shanghai [grant number. 15ZR1428900].
References
Wu J, Peters B A, Dominianni C et al. Cigarette smoking and the oral microbiome in a large study of American adults. Isme J 2016; 10: 2435–2446.
Lodi C S, Oliveira L V, Brighenti F L, Delbem A C, Martinhon C C . Effects of probiotic fermented milk on biofilms, oral microbiota, and enamel. Braz Oral Res 2015; 29: 01–07.
Lin S . Can the dental team shape dietary behaviour? BDJ Team 2016; 3: 19–20.
Buffie C G, Jarchum I, Equinda M et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 2012; 80: 62–73.
David L A, Materna A C, Friedman J et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol 2014; 15: 1–15.
Dominguez-Bello M, Costello E, Contreras M et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats. P Natl Acad Sci 2010; 107: 11971–11975.
Sampaio-Maia B, Caldas I M, Pereira M L, Pérez-Mongiovi D, Araujo R . The oral microbiome in health and its implication in oral and systemic diseases. Adv Appl Microbiol 2016; 97: 171–210.
Zaura E, Nicu E A, Krom B P, Keijser B J . Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol 2014; 4: 85.
Alshehri S S, Sweeney E L, Cowley D M et al. Deep sequencing of the 16S ribosomal RNA of the neonatal oral microbiome: a comparison of breast-fed and formula-fed infants. Sci Rep 2016; 6: 38309.
Dewhirst F E, Chen T, Izard J et al. The human oral microbiome. J Bacteriol 2010; 192: 5002–5017.
Gholizadeh P, Eslami H, Yousefi M, Asgharzadeh M, Aghazadeh M, Kafil H S . Role of oral microbiome on oral cancers, a review. Biomed Pharmacother 2016; 84: 552–558.
Zarco M F, Vess T J, Ginsburg G S . The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis 2012; 18: 109–120.
Zaura E, Keijser B J, Huse S M, Crielaard W . Defining the healthy 'core microbiome' of oral microbial communities. BMC Microbiol 2009; 9: 259–261.
Olsen I . The oral microbiome in health and disease. In Olsen I (ed). Oral infections and general health. pp 97–114. Springer International Publishing, 2016.
Camanocha A, Dewhirst F . Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J Oral Microbiol 2014; 10.3402/jom.v6.25468.
Lazarevic V, Whiteson K, Hernandez D, François P, Schrenzel J . Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 2010; 11: 523–534.
Crielaard W, Zaura E, Schuller A A, Huse S M, Montijn R C, Keijser B J . Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics 2011; 4: 1–13.
Papaioannou W, Gizani S, Haffajee A D, Quirynen M, Mamaihomata E, Papagiannoulis L . The microbiota on different oral surfaces in healthy children. Oral Microbiol Immunol 2009; 24: 183–189.
Kelvin L, Bihan M, Methé B A . Analyses of the stability and core taxonomic memberships of the human microbiome. Plos One 2013; 8: e63139–e63164.
Sonnenburg J L, Fischbach M A . Community health care: therapeutic opportunities in the human microbiome. Sci Transl Med 2011; 3: 78ps12–78ps21.
Belstrøm D, Holmstrup P, Nielsen C H et al. Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J Oral Microbiol 2014; 6: 1–9.
Illuzzi N, Galli R, Kushugulova A, Zhumadilov Z, Licciardello O, Marotta F . Expanding the Metchnikoff postulate: oral health is crucial in a successful global aging management strategy. Rejuvenation Res 2014; 17: 172–175.
Agbaje H O, Kolawole K A, Folayan M O et al. Digit sucking, age, gender and socioeconomic status as determinants of oral hygiene status and gingival health of children in suburban Nigeria. J Periodontol 2016; 87: 1047–1056.
Bhargava S, Motwani M B, Patni V M . Effect of handheld mobile phone use on parotid gland salivary flow rate and volume. Oral Surg Oral Med Oral Pathol Oral Radiol 2012; 114: 200–206.
Murshid E Z . Diet, oral hygiene practices and dental health in autistic children in Riyadh, Saudi Arabia. Oral Health Dent Manag 2014; 13: 91–96.
Tanner A C R, Mathney J M J, Kent R L et al. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol 2011; 49: 1464–1474.
Kato I, Vasquez A, Moyerbrailean G et al. Nutritional correlates of human oral microbiome. J Am Coll Nutr 2016: 1–11.
Adler C J, Malik R, Browne G V, Norris J M . Diet may influence the oral microbiome composition in cats. Microbiome 2016; 4: 23–32.
Jaiswal G R, Jain V K, Dhodapkar S V et al. Impact of bariatric surgery and diet modification on periodontal status: a six month cohort study. J Clin Diagn Res 2015; 9: ZC43–45.
Schutt C A, Neubauer P, Paskhover B, Fang-Yong L, Sasaki C T . The impact of dairy consumption on salivary inoculum. Dysphagia 2014; 29: 277–287.
Dagli N, Dagli R, Darwish S, Baroudi K . Oral microbial shift: factors affecting the microbiome and prevention of oral disease. J Contemp Dent Prac 2016; 17: 90–96.
Sapkota A R, Berger S, Vogel T M . Human pathogens abundant in the bacterial metagenome of cigarettes. Environ Health Perspect 2010; 118: 351–356.
Eaton T, Rd F J, von Reyn C F . Recovery of Mycobacterium avium from cigarettes. J Clin Microbiol 1995; 33: 2757–2758.
Kanwar A, Sah K, Grover N, Chandra S, Singh R R . Long term effect of tobacco on resting whole mouth salivary flow rate and pH: An institutional based comparative study. Eur J Gen Dent 2013; 2: 296–299.
Brook I . The impact of smoking on oral and nasopharyngeal bacterial flora. J Dent Res 2011; 90: 704–710.
Mason M R, Preshaw P M, Nagaraja H N, Dabdoub S M, Rahman A, Kumar P S . The subgingival microbiome of clinically healthy current and never smokers. Isme J 2015; 9: 268–272.
Moon J H, Lee J H, Lee J Y . Subgingival microbiome in smokers and non-smokers in Korean chronic periodontitis patients. Mol Oral Microbiol 2015; 30: 227–241.
Bizzarro S, Loos B G, Laine M L, Crielaard W, Zaura E . Subgingival microbiome in smokers and non-smokers in periodontitis: an exploratory study using traditional targeted techniques and a next-generation sequencing. J Clin Periodontol 2013; 40: 483–492.
Jabbour Z, Nascimento C . Assessing the oral microbiota of healthy and alcohol-treated rats using whole-genome DNA probes from human bacteria. Arch Oral Biol 2013; 58: 317–323.
Ahn J, Chen C Y, Hayes R B . Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control 2012; 23: 399–404.
Thomas A M, Gleber-Netto F O, Fernandes G R et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol 2014; 14: 1–12.
Wight A J, Ogden G R . Possible mechanisms by which alcohol may influence the development of oral cancer - a review. Oral Oncol 1998; 34: 441–447.
Barroso E, Martín V, Martínezcuesta M C, Peláez C, Requena T . Stability of saliva microbiota during moderate consumption of red wine. Arch Oral Biol 2015; 60: 1763–1768.
Daglia M, Papetti A, Grisoli P, Aceti C, Dacarro C, Gazzani G . Antibacterial activity of red and white wine against oral streptococci. J Agr Food Chem 2007; 55: 5038–5042.
Zaura E, Brandt B W, Mattos M J T D et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in faeces. Mbio 2015; 6: e01693–e01708.
Abeles S R, Jones M B, Santiago-Rodriguez T M et al. Microbial diversity in individuals and their household contacts following typical antibiotic courses. Microbiome 2016; 4: 39–51.
Ling Z, Liu X, Jia X et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep 2014; 4: 7485.
Ferrer M, Méndezgarcía C, Rojo D, Barbas C, Moya A . Antibiotic use and microbiome function. Biochem Pharmacol 2017; 134: 114–126.
Miller G E, Engen P A, Gillevet P M et al. Lower neighborhood socioeconomic status associated with reduced diversity of the colonic microbiota in healthy adults. Plos One 2016; 11: e0148952–e0148969.
Fujiwara N, Tsuruda K, Iwamoto Y et al. Significant increase of oral bacteria in the early pregnancy period in Japanese women. J Investig Clin Dent 2015; 8: e12189–e12197.
Adriaens L M, Alessandri R, Spörri S, Lang N P, Persson G R . Does pregnancy have an impact on the subgingival microbiota? J Periodontol 2009; 80: 72–81.
Jensen J, Liljemark W, Bloomquist C . The effect of female sex hormones on subgingival plaque. J Periodontol 1981; 52: 599–602.
Elattar T M . Prostaglandin E2 in human gingiva in health and disease and its stimulation by female sex steroids. Prostaglandins 1976; 11: 331–341.
Fujiwara N, Tsuruda K, Iwamoto Y et al. Significant increase of oral bacteria in the early pregnancy period in Japanese women. J Investig Clini Dent 2017; 8: 10.1111/jicd.12189. Epub 2015 Sep 8.
Gao S, Brown J, Wang H, Feng X . The role of glycogen synthase kinase 3-β in immunity and cell cycle: implications in esophageal cancer. Arch Immunol Ther Exp 2014; 62: 131–144.
Gao S, Li S, Ma Z et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer 2016; 11: 3–12.
Janati A I, Durand R, Karp I, Voyer R, Latulippe J F, Emami E . Association between oral conditions and colorectal cancer: A literature review and synthesis. Rev Epidemiol Sante Publique 2016; 64: 113–119.
Flynn K J, Baxter N T, Schloss P D . Metabolic and community synergy of oral bacteria in colorectal cancer. mSphere 2016; 1: e00102–e00116.
Nosho K, Sukawa Y, Adachi Y et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol 2016; 22: 557–566.
Momenheravi F, Babic A, Tworoger S S et al. Periodontal disease, tooth loss, and colorectal cancer risk: Results from the Nurses' Health Study. Int J Cancer 2016; 140: 646–652.
Gur C, Ibrahim Y, Isaacson B et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumours from immune cell attack. Immunity 2015; 42: 344–355.
Fan X, Alekseyenko A V, Jing W et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2016; 0: 1–8.
Huang J, Roosaar A, Axéll T, Ye W . A prospective cohort study on poor oral hygiene and pancreatic cancer risk. Int J Cancer 2016; 138: 340–347.
Curtis M A . Periodontal microbiology - the lid's off the box again. J Dent Res 2014; 93: 840–842.
Miyatani F, Kuriyama N, Watanabe I et al. Relationship between Cnm-positive Streptococcus mutans and cerebral microbleeds in humans. Oral Dis 2015; 21: 886–893.
Eberhard J, Stumpp N, Winkel A et al. Streptococcus mitis and Gemella haemolysans were simultaneously found in atherosclerotic and oral plaques of elderly without periodontitis—a pilot study. Clin Oral Investig 2016; 21: 447–452.
Huck O, Saadi-Thiers K, Tenenbaum H, Davideau J L, Romagna C, Laurent Y . Evaluating periodontal risk for patients at risk of or suffering from atherosclerosis: recent biological hypotheses and therapeutic consequences. Arch Cardiovasc Dis 2011; 104: 352–358.
Tonomura S, Ihara M, Kawano T et al. Intracerebral haemorrhage and deep microbleeds associated with cnm-positive Streptococcus mutans; a hospital cohort study. Sci Rep 2016; 6: 20074–20083.
Kocgozlu L, Elkaim R, Tenenbaum H, Werner S . Variable cell responses to P. gingivalis lipopolysaccharide. J Dent Res 2009; 88: 741–745.
Beck J, Garcia R, Heiss G, Vokonas P S, Offenbacher S . Periodontal disease and cardiovascular disease. J Periodontol 1996; 67: 1123–1137.
Blasi C . The autoimmune origin of atherosclerosis. Atherosclerosis 2008, 201: 17–32.
Scannapieco F A, Shay K . Oral health disparities in older adults: oral bacteria, inflammation, and aspiration pneumonia. Dent Clin North Am 2014; 58: 771–782.
Scales B S, Erb-Downward J R, Huffnagle I M, Lipuma J J, Huffnagle G B . Comparative genomics of Pseudomonas fluorescens subclade III strains from human lungs. BMC Genomics 2015; 16: 1032–1049.
Dickson R P, Erbdownward J R, Huffnagle G B . Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol 2015; 390: 1047–1055.
Dickson R P, Erbdownward J R, Martinez F J, Huffnagle G B . The microbiome and the respiratory tract. Physiology 2016; 78: 381–386.
Scannapieco F A . Pneumonia in nonambulatory patients: The role of oral bacteria and oral hygiene. J Am Dent Assoc 2006; 137: S21–S25.
Segal L N, Clemente J C, Tsay J C J et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 2016; 1: 16031.
Chhibbergoel J, Singhal V, Bhowmik D et al. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. Npj Biofilms Microbiomes 2016; 2: 7–20.
Mäntylä P, Buhlin K, Paju S et al. Subgingival Aggregatibacter actinomycetemcomitans associates with the risk of coronary artery disease. J Clin Periodontol 2013; 40: 583–590.
Ogrendik M . Rheumatoid arthritis is linked to oral bacteria: etiological association. Mod Rheumatol 2009; 19: 453–456.
Zhang X, Zhang D, Jia H et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015; 21: 895–905.
Whitaker T B . Standardisation of mycotoxin sampling procedures: an urgent necessity. Food Control 2003; 14: 233–237.
Xu X, He J, Xue J et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol 2015; 17: 699–710.
Luo W, Wen S, Yang L, Zheng G . Mucosal anti-caries DNA vaccine: a new approach to induce protective immunity against streptococcus mutans. Int J Exp Pathol 2017; 10: 853–857.
Hatta H, Tsuda K, Ozeki M et al. Passive Immunization against dental plaque formation in humans: effect of a mouth rinse containing egg yolk antibodies (IgY) specific to Streptococcus mutans. Caries Res 1997; 31: 268–274.
Al-Ghananeem A M, Leung K P, Faraj J, DeLuca P P . Development of a sustained antiplaque and antimicrobial chewing gum of a decapeptide. AAPS PharmSciTech 2017; 18: 2240–2247.
Khurshid Z, Najeeb S, Mali M et al. Histatin peptides: Pharmacological functions and their applications in dentistry. Saudi Pharm J 2016; 25: 25–31.
Presa M, Ortiz A Z, Garabatos N et al. Cholera toxin subunit B peptide fusion proteins reveal impaired oral tolerance induction in diabetes-prone but not in diabetes-resistant mice. Eur J Immunol 2013; 43: 2969–2979.
Zhang T, Wang Z, Hancock R E, De l F C, Haapasalo M . Treatment of oral biofilms by a D-Enantiomeric Peptide. Plos One 2016; 11: e0166997–e0167013.
Twetman S, Derawi B, Keller M, Ekstrand K, Yucellindberg T, Stecksenblicks C . Short-term effect of chewing gums containing probiotic Lactobacillus reuteri on the levels of inflammatory mediators in gingival crevicular fluid. Acta Odontol Scand 2009; 67: 19–24.
Zahradnik R T, Magnusson I, Walker C, Mcdonell E, Hillman C H, Hillman J D . Preliminary assessment of safety and effectiveness in humans of ProBiora 3 ™, a probiotic mouthwash. J Appl Microbiol 2009; 107: 682–690.
Di Pierro F, Donato G, Fomia F et al. Preliminary paediatric clinical evaluation of the oral probiotic Streptococcus salivarius K12 in preventing recurrent pharyngitis and/or tonsillitis caused by Streptococcus pyogenes and recurrent acute otitis media. Int J Gen Med 2012; 5: 991–997.
Ohshima T, Kojima Y, Seneviratne C J, Maeda N . Therapeutic application of synbiotics, a fusion of probiotics and prebiotics, and biogenics as a new concept for oral candida infections: a mini review. Front Microbiol 2016; 7: 10–18.
Petrou I, Heu R, Stranick M et al. A breakthrough therapy for dentin hypersensitivity: how dental products containing 8% arginine and calcium carbonate work to deliver effective relief of sensitive teeth. J Clin Dent 2009; 20: 23–31.
Que K, Fu Y, Lin L et al. Dentin hypersensitivity reduction of a new toothpaste containing 8.0% arginine and 1450 ppm fluoride: an 8-week clinical study on Chinese adults. Am J Dent 2010; 23 Spec No A: 28A–35A.
Schiff T, Delgado E, Zhang Y P, Cummins D, Devizio W, Mateo L R . Clinical evaluation of the efficacy of an in-office desensitizing paste containing 8% arginine and calcium carbonate in providing instant and lasting relief of dentin hypersensitivity. Am J Dent 2009; 22 Spec No A: 8A–16A.
Acknowledgements
The authors thank the National Science Foundation, China for financial support. Jia was responsible for writing. Zhi, Lai and Wang were responsible for modification and revision. Xia, Xiong and Zhang were responsible for language assistance. Che gave us the technology support in this paper. Ai was responsible for designing. All authors have read and approved the final article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jia, G., Zhi, A., Lai, P. et al. The oral microbiota – a mechanistic role for systemic diseases. Br Dent J 224, 447–455 (2018). https://doi.org/10.1038/sj.bdj.2018.217
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bdj.2018.217
This article is cited by
-
Isolation of phages against Streptococcus species in the oral cavity for potential control of dental diseases and associated systemic complications
Archives of Microbiology (2024)
-
The oral microbiome in autoimmune diseases: friend or foe?
Journal of Translational Medicine (2023)
-
The role of the oral microbiome in obesity and metabolic disease: potential systemic implications and effects on taste perception
Nutrition Journal (2023)
-
Multi-site microbiota alteration is a hallmark of kidney stone formation
Microbiome (2023)
-
Evaluation of the correlation between oral infections and systemic complications in kidney transplant patients: a retrospective pilot study
BMC Oral Health (2022)