Abstract
The human microbiome functions as an intricate and coordinated microbial network, residing throughout the mucosal surfaces of the skin, oral cavity, gastrointestinal tract, respiratory tract, and reproductive system. The oral microbiome encompasses a highly diverse microbiota, consisting of over 700 microorganisms, including bacteria, fungi, and viruses. As our understanding of the relationship between the oral microbiome and human health has evolved, we have identified a diverse array of oral and systemic diseases associated with this microbial community, including but not limited to caries, periodontal diseases, oral cancer, colorectal cancer, pancreatic cancer, and inflammatory bowel syndrome. The potential predictive relationship between the oral microbiota and these human diseases suggests that the oral cavity is an ideal site for disease diagnosis and development of rapid point-of-care tests. The oral cavity is easily accessible with a non-invasive collection of biological samples. We can envision a future where early life salivary diagnostic tools will be used to predict and prevent future disease via analyzing and shaping the infant’s oral microbiome. In this review, we present evidence for the establishment of the oral microbiome during early childhood, the capability of using childhood oral microbiome to predict future oral and systemic diseases, and the limitations of the current evidence.
Similar content being viewed by others
Introduction
Human life is dependent on a diverse community of symbiotic microbiota that has co-evolved with their human host. The interactions between microbiota and host modulate crucial aspects of the host’s normal physiology, metabolism, immunity, and neurologic function.1 In the adult population, the host-microbiome at various body sites has reported associations with systemic diseases. For instance, dysbiosis in the gut microbiome could induce an ecological shift of the microbial community, from a physiological to a pathological composition, lead to dysregulated production of harmful microbial-derived products or metabolites, and contribute to or become a risk marker for a diverse range of local and systemic diseases. The reported diseases that have a gut microbial association include but not limited to inflammatory bowel diseases.2,3 celiac disease (CD),4,5 depression.6and Alzheimer’s disease.7 Intriguingly, an intricate linkage between the dysregulated gut microbiome and osteoarthritis was recently revealed. The loss of beneficial Bifidobacteria while an increased abundance of key proinflammatory species in the gut microbiome is found to be associated with obesity; this microbial imbalance event is further linked with a downstream of systematic inflammation that could accelerate knee osteoarthritis.8 Associations between oral microorganisms and cancers (oral and esophagus,9,10,11,12 pancreatic,13 and colorectal14) have also been suggested recently. Moreover, periodontal microorganisms, i.e., Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, Provetella intermedia, have been found in human atheromatous plaques at various sites, with implied association with vascular disease.15 Furthermore, a dysbiosis in the vaginal microbiome is associated with a variety of adverse outcomes, including miscarriage, invasive maternal and neonatal infections, and preterm birth delivery.16,17,18,19,20
Through the first years of life, the newborn infant microbiota is highly dynamic and undergoes rapid changes in composition, towards a stable adult-like structure that harbors distinct microbial communities of unique composition and functions at specific body sites.21,22,23,24,25,26,27,28 Colonization of oral mucosal surfaces begins at birth with the introduction of bacteria and fungi through multiple paths, including maternal transmission during childbirth, parental exposures, diet and horizontal transmission from caregivers and peers.29,30,31,32 The oral microbial community continues to develop with the eruption of primary teeth in early infancy and establishment of permanent dentition in children, evolves into a complex and diverse microbiome.33 A complex interplay between establishment and development of the neonate’s immunity and early microbial acquisition occurs.34,35 These early life interactions between the microbiome and human host are responsible for features of postnatal innate and acquired immune functions and physiological development that influence future health.28,36,37,38
The oral cavity serves an initial entry point for colonization of the oral and gut microbiota39,40 and therefore is an easily accessed body site for assessment of the microbial community, and biologic markers used to diagnose, predict, and monitor both oral and systemic diseases.41 Similar to reported associations between microbiome and adults’ health, recent data suggest that disruptions in early oral colonization and establishment of a healthy oral microbiome may influence the progression of both oral and systemic conditions in children. Despite that more longitudinal studies are critically needed to provide substantial evidence on causal relationship between the oral microbiome and oral/overall health, health conditions that potentially have an oral microbial involvement and harbor oral microbial signatures include but not limited to children’s tooth decay,42,43,44,45,46,47,48 infant weight gain,40 pediatric appendicitis,49 and pediatric inflammatory bowel disease.50
To the vulnerable populations, infants and young children, oral sample collection in forms of saliva and mucosal swabs is noninvasive and thus presents an optimal diagnostic medium that holds great promise for use as diagnostic tools. Moreover, researchers are searching for the potential utility of the microbiome with a particular focus on manipulating microbials.51 Microbial intervention of a single bacterial strain was demonstrated to be effective in altering disease risk in the low-complexity microbiome of the newborn gut.52 Although, to date, no studies have addressed the impact of manipulation of the oral microbiome on systemic disease, as the microbiome research moves on a fast track, we can envision a future where early life salivary diagnostic tools could be used to understand better and shape our oral and systemic health. Therefore, this review attempts to gather evidence and elucidate the establishment of the oral microbiome during early childhood, the factors associated with oral microbial profiling, and the capability and challenges of using childhood oral microbiome to predict future oral and systemic diseases.
Oral microbial community establishment in early childhood
Several terms are used to define different stages of early childhood. Medically, the newborn or neonate is defined as an infant in the first 28 days after birth. The term “infant” typically refers to young children under 1 year of age, or some definitions expand this period to up to 2 years of age. When a child learns to walk from age 1 to 4, the term “toddler” may be used instead. The period of birth to preschool age (5–6 years old) is considered as early childhood.
Oral microbial colonization is traditionally considered to take place after birth; however, recent studies have brought our attention to the commence of the human microbiome before birth. Studies reported the presence of microorganisms in amniotic fluid in up to 70% of the pregnant women, and particularly the presence of several oral microorganisms, such as Streptococcus, Fusobacterium, Neisseria, Prevotella, and Porphyromonas, in the human placenta.53,54 To assess the possible origin of the placental microbiome, Gomez-Arango et al.55 examined the gut, oral, and the placental microbiome from pregnant women using 16S rRNA sequencing. Three shared genera, including Prevotella, Streptococcus, and Veillonella, were found in all gut, oral, and placenta samples.55 Surprisingly, the placenta microbiome did not harbor unique core genera when the microorganisms residing in the gut and oral cavity were compared.55 Moreover, although the placental colonization may have multiple sources (oral and gut), the placental microbiome resembles more like the pregnant oral microbiome.55
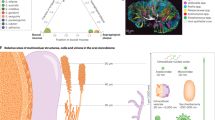
During and after birth, the newborn exposes to a wide variety of microorganisms, e.g., bacteria, fungi, parasites, and virus, under the influence of contact routes and an infant’s immune tolerance.56 Remarkably, the oral microbial colonization follows a temporal and spatial sequential, and only a subgroup of microorganisms become permanent residents of the oral cavity,57 illustrated in Fig. 1. The set of earlier colonizers seems to condition the subsequent colonization, which leads to more complex and stable ecosystems in adulthood.58
Oral bacterial community
Immediately after birth, with contact with the outside world through breathing, feeding, and the contact with care providers, the colonization and establishment of microbial pioneers in the oral cavity take the stage. In a cross-sectional designed study, Mason et al.29 analyzed the oral mucosal swab samples from 47 infants at the prdentate stage and identified 178 species-level operational taxonomic units (s-OTUs) that belong to 50 genera, with (65 ± 18) s-OTUs in each infant. The majority of these identified species were Gram-positive facultative, followed by Gram-negative anaerobes..29 Mason et al.29 further identified a core of oral microbiome for predentate infants, including genera Streptococcus, Gemella, Granulicatella, and Veillonella. Interestingly, these core species only accounted for 45% (23%–61%) of the total oral microbiota of each infant.29 Out of the 178 identified species, only 33 were shared by ≥75% of infants. The mean number of shared core species by each infant was 27 ± 5.29
As an infant grows, oral bacterial diversity and richness continue to increase through time. Through sequencing of 16S rRNA gene V3–V4 hypervariable regions, Dzidic et al.59 examined the temporal evolution and maturation of the oral microbial ecosystem in early infancy and childhood using a longitudinally collected oral sample set from 90 children. These children were followed from birth to 7 years of age. Before tooth eruption, the mucosal surfaces serve as primary sites for bacterial colonization. The most frequently detected early colonizers in the oral cavity are Streptococcus (Streptococcus epidermidis and Streptococcus salivarius), Staphylococcus spp., and Fusobacterium..29,59 The reasons for the high abundance of Streptococcus in the early oral cavity lie in: (a) Streptococcus spp. are capable of adhering to epithelial cells and (b) Streptococcus spp. are one of the dominant bacterial groups found in human breastmilk; the oral settlement of Streptococcus spp is initiated by direct transmission via physical contact through bread feeding, facilitated by an appropriate nutrient supply in breast milk that favors the growth of Streptococcus spp, and further reinforced by repeated breast milk feeding.60 Among Streptococcus species, Streptococcus salivarius is the most often found species in the oral cavity of the newborn61,62, its abundance reaches the highest at 3 months of age, accounting for 10%–15% of the total Streptococcal species. The abundance of S. salivarius decreases steadily its peak at 3 months, likely opposing to teeth eruption.30 In addition to the early colonizers, bacterial genera Gemella, Granulicatella, Haemophilus, and Rothia present at 3–6 months of age with more than 1% of abundance; and their abundance further increases with time.59
Remarkably, the oral ecosystem reforms with the eruption of the first tooth. The salivary microbial community of the primary dentition reveals a significant greater alpha diversity and equitability when compared to that of the predentate infants.29 A lower level of Gram-positive facultative and a higher level of Gram-negative facultative are seen when transitioning from the predentate to primary dentition.29 For instance, in the first several months of an infant’s life, before tooth eruption, Escherichia coli, Pseudomonas, Staphylococcus, along with the lactic acid-producing bacteria such as Lactobacillus gasseri, Lactobacillus crispatus, and Streptococcus spp are highly prevalent.61,63,64 Along with the tooth eruption providing new binding sites, new ecological events take place in the oral environment. Streptococcus mutans, for example, accelerates its colonization at this stage, due to the emergency of their preferable adhesion surface, teeth. As time goes by, Fusobacteria, Synergistetes, Tenericutes, TM7, and SR1 dominate the oral microbial community by the end of first year of life.65 Post tooth eruption, although most oral microorganisms colonize all oral cavity sites, including mucosa, tongue, and teeth; their proportion may differ depending on the colonization sites. A higher microbial load is found on the teeth and tongue comparing to the oral mucosa and saliva. In parallel to developing specific microbial niche at different oral sites, the sral bacterial diversity and richness continue to increase through time. At the end of primary dentition, 7 years of age, nearly 550 OTUs were identified in the oral cavity; the Shannon diversity index reaches approximately 2.4 (ref. 59).
After tooth eruption, the plaque microbiota forms a new, distinct oral community. As noted in our study, while comparing the oral microbial community between preschool children with and without caries using 16s sequencing, the microbial composition differed notably between saliva and plaque samples, regardless of caries status.48 At the genus level, the two most abundant genera in children’s saliva were Streptococcus and Veillonella, whereas, in plaque, the most abundant genera were Veillonella, Streptococcus, Actinomyces, Selenomonas, and Leptotrichia.48 At the species level, three dominant taxa represented 60% of the relative abundance in the salivary community: Streptococcus ET_G_4d04, Veillonella atypica_dispar_parvula, and Streptococcus vestibularis_salivarius.48 In contrast, the five most abundant species, representing 30%–50% of the bacterial plaque community, were Veillonella atypica_dispar_parvula, S. mutans, S. ET_G_4d04, Streptococcus oral_taxon_B66, and Streptococcus gordonii.4
Oral fungal community
In addition to bacterial community, the oral cavity of newborns is also known to be colonized by fungal community, specifically Candida, starting on the first day of life; during the first year, the rate of oral colonization by Candida may vary, depending on the study population and detection methods, the detection rate range between 40% and 82%.66,67,68 For an extended period, Candida was the only fungus recognized as part of the normal oral microbial population, despite its opportunistic character.69 In 2010, a metagenomic study identified 74 genera of cultivable fungi and 11 uncultivable fungi in the oral cavity of healthy adults.70 Although Candida was the most frequent genus isolated in 75% of the subjects, other fungi groups presented a relevant prevalence, such as Cladosporium (65%), Aureobasidium (50%), Saccharomycetales (50%), Aspergillus (35%), Fusarium (30%), and Cryptococcus (20%).70
Most recently, the skin, oral, and anal mycobiomes were analyzed by Ward et al.32 among an infant cohort from birth through 30 days of life, using amplicon sequencing of internal transcribed spacer 2 (ITS2). Ward et al.32 noted that between colonization sites, the alpha diversity of the oral mycobiome was significantly lower than that of skin and anal mycobiomes. Within individuals, the infant oral mycobiomes exhibited high intra-individual variability for beta diversities over time; however, when measured by weighted UniFrac distances, no distinct clusters were identified.32 Mycobial community differs across the body sites. On infant skin, the most abundant and prevalent mycobial taxa were C. tropicalis, C. parapsilosis, S. cerevisiae, C. albicans, and C. orthopsilosis; in infant oral cavity, the abundant and prevalent taxa were C. parapsilosis, C. tropicalis, S. cerevisiae, C. orthopsilosis, C. albicans, and Cladosporium velox; among the infant anal mycobiome, the most abundant taxa were C. parapsilosis, C. tropicalis, C. albicans, S. cerevisiae, C. orthopsilosis, and Cryptococcus pseudolongus.
Nevertheless, since there have been few reported longitudinal studies of mycobial community in early infancy, the role of this oral “mycobiome” and their identification in the oral cavity of children remains underexploited.
Oral viral community
Viruses have also been found in the oral cavity, with their presence primarily viewed as a pathological nature. In the past decade, there have been only a handful of publications reporting the oral virome, which is in contrast to the oral microbiome, an area that demands critical attention. The oral cavity harbors 300–2 000 viral genotypes in any individual.71 The most common viruses isolated from the oral cavity include rotavirus, norovirus, HIV, hepatitis C virus, herpes simplex viruses 1 (HSV1) and HSV2, Epstein–Barr virus and influenza viruses.72 In addition to these commonly reported virus, some less abundant viruses, e.g. eukaryotic DNA viruses including herpesvirus HPV7 and Anelloviruses, as well as some RNA viruses, are also permanent constituents of human mouth.72,73
Several viral agents can infect oral mucosal cells; however, only a few cause clinical manifestations. In children, the severity of symptoms is related to the age at which the infection was acquired. Newborns could be infected by HSV1 and HSV2, which can cause herpetic gingivostomatitis, orofacial herpes, and aphthous stomatitis.74 Neonatal HSV infection is uncommon, with the reported incidence rates as 1.6–33 per 100 000 live births.75 HSV infection in a newborn can be very severe and even cause death due to the compromised and underdeveloped infant immune systems.76 Other viruses that could colonize in the oral cavity during early childhood are the Coxsackie A virus, which causes herpangina and hand, foot, and mouth disease; the Morbillivirus that causes measles; the Rubulavirus that causes mumps; and the human papillomavirus that causes oral papilloma (warts).77
Factors shaping children’s oral microbiome
For healthy adults, an individual’s oral microbiome is associated with many factors, including the time of the sample collection, age, gender, diet, extreme environment, etc.78. Similarly, the host and environmental factors also influence the assembly of the oral microbiome in early childhood. The most studied factors include genetics, terms of labor, delivery mode, antibiotics use during birth and early infancy, feeding method, and maternal oral microbiome characteristics. These influencing factors contribute to shaping both bacterial and fungal communities.78,79 The summary in Fig. 1 highlights the factors that shape children’s oral microbiome.
Genetic determinants
Even though few studies have identified genes that contribute to microorganism infections,80 the influence of human genes on the colonization and establishment of the oral microbiome remains under-investigated. Demmitt et al.81 examined the contributions of host genetics and environmental factors to salivary microbiome composition using an unbiased genome-wide association study (GWAS) analysis in 752 twin pairs from the Colorado Twin Registry. A method that used KGG4 that sums single-nucleotide polymorphisms (SNP) significance across coding genes were used. Several loci reached genome-wide significance after correcting for multiple testing, including one locus on chromosome 7 near the gene IMMPL2 and one locus on chromosome 12 near the non-coding RNA gene INHBA-AS1 (ref. 81).
Exploring the effects of host genotype on the oral microbiome composition and caries phenotypes also gained attention. While several heritable oral taxa were identified, the scientific finding has not reached agreement on whether the abundance of traditionally considered cariogenic bacteria hold inheritable traits. Gomez et al.82 profiled the supragingival plaque microbiome from 485 dizygotic and monozygotic twins, with a mean age of (7.8 ± 1.4) years old. This research group revealed that the similarity of the oral microbiota increased with a shared host genotype, regardless of caries status. Interestingly, the most heritable oral bacteria, such as P. pallens, were not associated with caries status but associated with age and sugar consumption. P. pallens decreased in abundance when children are older and sugar consumption frequency increase. However, the results from Gomez et al. stating that some the abundance of certain cariogenic bacteria are lack of heritable traits are in contrary to some other studies. Previous twin studies used culture- and array-based approaches to obtain microbial information and examined the heritability of oral microbes. These studies resulted indicated that the abundance of the cariogenic S. mutans and specific pathogenic attributes such as acid production in dental plaque and saliva have a close genetic correlation.83,84 Furthermore, the abundances of Prevotella, Pasteurellaceae, and Leptotrichia have been previously found to be associated with SNPs in host genes that code for ATP-binding cassettes, protein synthesis, cell division, and tumor suppression.83
Human genetics also influence individual’s susceptibility to fungal infection. Abusleme et al.85 demonstrated that individuals with defects in STAT3 are prone to the dysbiosis of oral fungal and bacterial community, and susceptible to recurrent oral fungal infections. STAT3 is a protein coding gene, playing a critical role as transcription factors in the downstream of cytokine signaling from interleukin-6 (IL-6), IL-21, IL-10, and IL-23—a group of human immune mediators. Patients with autosomal-dominant hyper-IgE syndrome (AD-HIES) harbor loss-of-function (LOF) mutations in the STAT3 gene.86,87 In turn, a severe dysbiosis of fungal community was found in AD-HIES patients’ oral mucosa, with a dominance of C. albicans and minimal representation of health-associated fungi.85 The STAT3 defect in participants with AD-HIES was also associated with distinctive bacterial communities, particularly with a reduced microbial diversity and an enrichment of Streptococcus oralis and S. mutans. Moreover, increased abundance of cariogenic bacteria—S. mutans—was consistent with the increased risk for dental caries among patients AD-HIES.85
Preterm and full-term pregnancy
In medical context, preterm refers to the condition that infants born alive before 37 weeks of pregnancy.88 Sub-categories of preterm birth, based on gestational age, include extremely preterm (<28 weeks), very preterm (28–32 weeks), and moderate to late preterm (32–37 weeks).88 Globally, approximate 15 million babies are born preterm yearly.89
Although a small number of studies have compared the oral microbiota among infants with different pregnancy terms, difference in the oral microbiota diversities between the full-term and preterm infants was not discovered. For instance, in a cross-sectional designed study, Younge et al.90 characterized the oral and skin microbiota of 89 preterm (23–36 weeks) infants in intensive care unit with an average age of 42 days (1–252), and 40 full-term infants with an average age of 1 day (0–122) during their birth hospitalization. Among the preterm infants, 48% were white, 50% were Black or African American, and 3% were unknown races; among the full-term infants, 51% were white, 31% were Black or African American, and 15% were unknown races.90 The samples collected from the oral cavity and various skins sites included the forehead, posterior auricular scalp, periumbilical region, inguinal folds, and upper thighs.90 Among the oral microbiota of full-term and preterm infants, no significant difference were found in alpha diversity measured by the Shannon diversity index.90 The beta-diversity measured by principal coordinate analysis differed significantly between the oral and stool samples (P < 0.000 1), and between the skin of the upper body and oral samples (P = 0.004 9).90 In contrast to the oral community, the skin bacterial community differed significantly by gestational and postnatal age, and body sites.90 Another small-scale study analyzed the oral microbiota of seven preterm infants (25–27 weeks gestational age) at three timepoints in the first five days after birth. This study revealed a transition of the predominate species in the infants’ oral cavity, from a predominance of Mycoplasmataceae and Moraxellaceae in infants’ first 36 h of life to a predominance of Staphylococcaceae and Planococcaceae by day 5 (ref. 91).
Concerning enough, when a comparison was made between the full-term and preterm infants with extremely low birth weight (LBW), who was less than 100 0 g in the first 6 weeks of life, preterm with extremely LBW infants were prone to be colonized with pathogenic microorganisms in the oral cavity.92 Approximately 20% of the preterm and extreme LBW infants by week 1 and over 50% of the preterm/extreme LBW infants by week 6 were colonized with pathogens, e.g., methicillin-resistant Staphylococcus aureus (MRSA).92
A critical point to be addressed is while the infants born full-term have ongoing interaction with the mother, family, and home environment, which might affect the early oral microbiome development. Unfortunately, preterm or LBW infants are usually separated from their mothers immediately after birth and are cared by medical personnel in the complex environment of the neonatal intensive care unit (NICU) for a period ranging from days to months. Given NICU has its unique environmental microbes, including commensal and pathogenic organisms,93,94 the infant’s ongoing and exclusive interaction with medical personnel, in addition to receiving specific medical procedures and medications in NICU, has the potential to impact preterm and LBW infants’ oral microbial colonization.95
Moreover, antibiotic use in NICUs remains widely diverse and controversial. Antibiotics are effective in treating pathogenic bacteria-related infections but simultaneously deplete commensal bacteria and promote the growth of opportunistic fungal species. Studies have shown the association between neonatal antibiotics use and decreased levels of Clostridial colonization in infants’ gut when administered in a prolonged period.96 Intriguingly, a study by Gomez-Arango et al.97 indicated that maternal antibiotics use during delivery also impacts the infant oral microbiome development. In this study where the infant oral swabs were sampled in the first three days after delivery, the study results showed that Proteobacteria were abundant in infants with maternal intrapartum antibiotics exposure while Streptococcaceae, Gemellaceae, and Lactobacillales were dominant in unexposed neonates.97
Delivery mode
Similar to what occurs to gut microbiota in early infancy, some studies found that the delivery modes—vaginal delivery or cesarean section—also influence the oral microbial community development in infants; however, the term of influence remains unclear. Immediately after birth, the bacterial niche in different habitats of the newborn (oral, nasopharyngeal, skin, and intestines) are very similar to each other.23 Nevertheless, as time goes by, bacterial communities colonized on infants born vaginally resemble mothers’ vaginal bacterial communities, predominantly Lactobacillus, Prevotella, Bacteroids, TM7, and Sneathia spp., whereas the bacterial profile of infants born by cesarean section resemble those present in mothers’ skin, predominantly Staphylococcus, Corynebacterium, Slackia, Veillonella, and Propionibacterium spp.23,98,99
A birth cohort study conducted in Ireland among 84 infants from birth to the first year of life revealed that birth mode did pose an impact on the infant microbiota, but only up to the first week of age.100 The influence disappeared beyond the first week after birth.100 Within the first week, higher bacterial community diversity measured by Shannon index (P = 0.037) and PerMANOVA assay (P = 0.047) was detected in the C-section group than those in the vaginally delivered infants.100 On the contrast, a Sweden birth cohort of 59 infants did not reveal associations between birth route and young children’s oral microbiome.101 The difference in the bacterial community in early infancy that associated with birth route might remain or reduce, or might even have long-term effects on childhood oral microbiome composition, metabolism, and implications to overall health.102 However, these projections still need to confirmed by more well-designed longitudinal birth studies.
Feeding method
Similar to the influence of feeding methods on the gut microbiota,103 the oral microbiota of formula-fed and breastfed infants have distinct characteirstics.104 The difference in infants’ oral microbiome may arise due to the milk-related bacteria transmission,105 effects of various milk components on the attachment of bacteria in the oral cavity,106 differed utilization of carbohydrates in breastmilk and formula by bacteria, and/or infant’s boosted innate immunity from breast milk and its effect on early oral microorganisms colonization.107
Researchers from Sweden108 characterized and compared the oral microbiome in formula-fed and breastfed infants. The oral microbiota pattern of breastfed infants differed markedly from the formula-fed infants, with significantly lower species richness at 4 months of age.108 However, notable enough, this difference in oral species richness between breastfed and formula-fed infants disappeared when these infants reached 12 months of age. In contrast to the species richness, the difference of certain microbial community characteristics remained even after the discontinuation of the breastfeeding, which indicates that there might be a long-term effect of breastfeeding on the oral microbiota and this phenomenon deserves further follow-up.
Moreover, this study examined how types of formula could influence the oral microbiota development. An experimental formula that was supplement with bovine milk fat globule membranes was compared to standard formula. Researchers found that although oral species richness did not differ between the experimental and standard formula groups, consumption of the experimental formula was associated with significant differences in taxa abundance, e.g., lower level of Moraxella catarrhalis found in the experimental formula group. Furthermore, when infants reached 2 years of age, oral bacterial diversity appears to be higher in children who abandoned breastfeeding before turned 1 year of age.59
Regarding S. mutans, a bacterial species that is famous for its acidogenicity, aciduricity, and capability of synthesizing extracellular matrix using carbohydrates, although not conclusive, some studies demonstrated that when the presence of S. mutans in the oral cavity of 1-year old infants was more prevalent in formula-fed than in breastfed infants.109 Previously, in vitro studies revealed that both human and bovine milk may inhibit the metabolism and adhesion of S. mutans.110 Although the association between breastfeeding and risk of caries is debatable since S. mutans is a known culprit for caries, the supporting evidence that formula-fed infants harbor more S. mutans at 12 months of age indeed support the beneficial role of breastfeeding in preventing dental caries.111. In addition to breastfeeding and bottle feeding, introducing comforting oral device, such as pacifier, has been found to serve as a potential risk factor for the colonization of S. mutans in infant’s mouth in early infancy.112
When considering the influence of feeding methods on infant oral microbiome, one limitation to be underlined is study groups from reported literatures were often not randomized, which might pose unknown group differences correlated to oral microbiota acquisition, in addition to the feeding methods. As a commonly limitation for non-randomized trials, these imbalanced factors between groups, e.g. race, ethnicity, age, gender, health conditions, caregivers’ oral and overall health, are confounding factors and could lead to biased results.
Maternal influence
Colonization of oral mucosal surfaces begins at birth with the introduction of bacteria and fungi through a variety of paths, including maternal perineum–infant oral transmission during childbirth,23 parental exposures, digit sucking, diet, and horizontal transmission from caregivers and peers.29,30,31 Strain-resolved metagenomic profiling confirms transient maternal transmission of strains originating from multiple sources, with the vaginal, skin, oral, and gut communities all contributing to the early infant microbiome.113 However, a few days after birth, the contribution from the vaginal and skin microbiome have decreased, while gut microbes from the mom persist in the infant’s gut presumably through oral entry.113 By day 3 of life, 95% of the infant’s oral microbiome was shared with maternal microbiomes.113 Although evidence supports the maternal–neonate transmission of pathogenic microorganisms, e.g., Candida species immediately after birth into early infancy,114,115 more importantly, the maternal–neonatal transmission of microbes also serves an evolutionary survival function by preparing the infant to adapt to the environment by prepping the sterile infant gut with maternal microbes.116,117 Acquisition of maternal microbes gives the infant gut a head start through colonization with favorable symbiotic microbes, potentially protecting the infant from life-threatening respiratory and diarrheal infections that were a major cause of infant mortality in the early twentieth century and remain an important cause in some developing countries.
As a significant number of the human-body early colonizer are of maternal origin, we have recently compared the oral microbiota in a set of mother and preschool children, in which the children were grouped, based on their caries status, into severe early childhood caries (ECC) or caries-free. We observed that the microbial community structure and diversity within the mother–child dyads were similar (although species abundance differed across pairs) despite the caries status. Our observed strong maternal influence on the oral microbiota of children is supported by the recent observations of Chu et al.118, who found that the infant and maternal oral microbiota shared a similar community structure and taxonomic membership. Another study by Mason et al. revealed that 85% of infants’ oral microbiota resemble their mothers’ in the first six months after birth, suggesting maternal oral microbiota plays a role in seeding microbial species to the child. However, intriguingly, the mother–child oral microbiome similarity significantly decreased with the eruption of primary teeth in a child’s mouth, often, around 6–8 months.29
Similar to the bacterial community, maternal prints have been found in the infant oral mycobiomes.32 Infant and maternal oral mycobiomes were similar to each other when alpha and beta diversities were compared; the total numbers of observed taxa were comparable in infant and adult samples for throughout longitudinal observational points, regardless of body site (oral, skin, and anal).32 Among fungal species present in the oral cavity, C. albicans is the most prevalent species. C. albicans colonizes in the oral cavity of neonates as well, both vertical and horizontal transmission was implied to their emergence. The vertical transmission ranges from 14% to 41% when measured by different methods, e.g., electrophoretic karyotyping, restriction endonuclease analysis of genomic DNA, and DNA fingerprinting.119,120 Our study examined the relatedness of C. albicans among 18 preschool children and mother dyads using restriction endonuclease analysis of the C. albicans genome and found more than 60% of the child–mother dyads harbored identical C. albicans strains.121 While no caspofungin-resistant strains were identified when compared to the wild type, C. albicans isolated from more than 65% of child–mother dyads demonstrated similar susceptibility to the antifungal medication caspofungin.121
Health implications in children’s oral microbiome
The oral microbiome remains its stability over time in healthy individuals, despite subjected to a variety of host and environmental challenges. The distinct oral microbial community is associated with a series of oral and systematic diseases.10,122,123,124,125,126,127,128 Although a majority of these studies are cross-sectional or case–control designed, with small sample sets which are incapable of establishing causative relationship between oral microbiome and diseases, changes in the characteristics of the oral microbiota may provide correlative insight and projection into the onset, progression, and recurrence of human diseases. Here, we further zoomed into the children’s health and summarized studies that have provided critical insights into the oral microbial markers of the oral and systemic health in children. The oral taxonomic characteristics associated with each disease status are summarized in Table 1.
Oral microbiome and early childhood caries (ECC)
Although largely preventable, ECC remains the single most common chronic childhood disease, with nearly 1.8 billion new cases per year globally.129,130 The microbial etiology of ECC is linked with poly-bacterial infection of teeth. Traditionally, S. mutans is considered as a prime culprit for ECC due to its acidogenicity, aciduricity, and ability to form extracellular glucans.131,132,133,134,135,136,137 Although at very low levels, S. mutans was detected in the oral cavity of the infants in early infancy, even before tooth eruption;138 and with a trend of the increasing amount with the presence of teeth and notably higher in children with ECC.59 Interestingly, a recent analysis of calcified dental plaque shows that the composition of oral microbiota remained relatively constant between Neolithic and Medieval eras. Cariogenic bacteria (e.g., S. mutans) became dominant beginning with a major production of processed flour and sugar, beginning with the industrial revolution.139 Well-designed longitudinal studies have demonstrated the predictive power of using S. mutans to predict ECC risk. Fontana et al.140 prospectively followed a cohort of 329 US children ((26 ± 6) months old) for 1 year and reported that children with more than 105 colony-forming unit per mL salivary S. mutans at the baseline were at higher risk for developing ECC. Piva et al.141 evaluated a cohort of 163 Brazilian children (3–4 years old) in a 2-year prospective period, and found higher S. mutans counts were associated with caries progression.
In addition to S. mutans, several studies have characterized the oral microbiota in caries-active children, and have identified additional species that are associated with caries state, including S. salivarius, S. sobrinus, S. parasanguinis, S. wiggsiae, S. exigua, L. salivarius, Parascardovai denticolens, Porphyromonas, Actinomyces, and Veillonella.42,43,44,45,46,47 Gross et al.42 sequenced the plaque samples from 36 severe ECC (S-ECC) and 36 caries-free (CF) children with a mean age of 23.6 months and monitored the microbiota evolve during the onset of S-ECC. They confirmed that S. mutans was the dominant species in many, but not all children with S-ECC.42 Other species that had elevated quantities among children with S-ECC were S. salivarius, S. sobrinus, and S. parasanguinis. Elevated levels of these species were observed specifically among children with no or low levels of S. mutans, suggesting these species were playing alternative pathogenic roles, and that targeting species in addition to S. mutans may be promising interventions for ECC and S-ECC.42 Among children without past caries history, Veillonella, not S. mutans or other acid-producing species, were found to be a predictor for future caries.42 The levels of Veillonella highly correlated with total acid-producing species.42 The underline explanation of the phenome lies in that Veillonella is well-known for metabolizing lactate; lactate, in turn, is an end-product from the carbohydrates-derived catabolism by Streptococcus species that many of them are associated with caries. An inspiring way to elaborate is that Veillonella might not be acting as a criminal for causing caries, but a “whistleblower” for caries. On the contrary, with the occurrence of caries and advancement of caries stages, the abundance of specific taxa reduced, for instance, Streptococcus mitis group, Neisseria, and S. sanguinis.42 In concert with the different abundance of cariogenic and symbiotic bacteria in caries and healthy children, community diversity was also reduced in children with caries as compared to their healthy counterparts.42
Intriguingly, available microbiological data also show that this pediatric oral disease, ECC/S-ECC, has a strong fungal involvement. Particularly, Candida species (especially C. albicans) are more prevalent in the oral cavity of children with ECC or S-ECC (over 80%) in comparison to caries free children (approximately 20%), and a higher abundance in the ECC/S-ECC group; the carriage of the fungus is positively correlated with the carriage of S. mutans and the severity of dental caries in children, as reviewed by Xiao et al.142. The presence of C. albicans in preschool children’s oral cavity is associated with a 6.7 times higher risk of experiencing ECC than those ones do not have this fungal species.142 Our recent findings48 demonstrated that the presence of C. albicans is further associated with alterations of oral bacterial composition and cariogenic bacteria virulence that could contribute to an oral environment that are more prone to caries. The C. albicans-associated oral bacteriome is characterized as an enrichment of a highly acidogenic–aciduric bacterial community, with an increased abundance of plaque Streptococcus (particularly S. mutans), certain species of Lactobacillus/Scardovia and salivary/plaque Veillonella and Prevotella, and decreased levels of salivary/plaque Actinomyces.48 Following the enrichment of S. mutans in plaque, the enzymatic activity of glucosyltransferases, key enzymes that could synthesize extracellular matrix by S. mutans, was also enhanced. Our finding from the clinical setting was verified in rodent models by Bowen and Koo,143 indicating that C. albicans could play an essential role in enhancing plaque glucosyltransferase enzymatic activities and caries pathogenesis. Therefore, in addition to cariogenic bacteriome, the need for including fungal counterparts in oral microbiome research in the context of ECC/S-ECC, and understanding its role in the onset, progression, and relapse of this disease in future longitudinal studies are paramount, by doing so could potentially lead the way to prevent and treat this costly and intractable disease from an innovative fungal perspective.
Besides cariogenic bacteria and yeast, S. sanguinis has received much attention in the context of caries prevention. S. sanguinis starts its oral colonization in the second half of the first year of infants, in association to tooth emergence,144 following a similar pattern of development regardless of children’s caries experience.59 Researchers believe S. sanguinis could be playing a benign role in the oral cavity, and have an antagonistic role in dental caries since it may compete with cariogenic S. mutans for colonization sites on tooth surface.144
Association between children’s oral microbiome and systematic diseases
Infant oral microbiome and weight gain
The oral microbiome may play a vital role and serve as an indicator of infant development. A recent study40 revealed that oral microbiome harbors a better bacterial signature for predicting the weight gain in the early infancy when compared to the gut microbiome. In this study, via 16S rRNA sequencing, the authors analyzed the gut and oral microbiota of 226 young children at seven times points in the first two years of life, along with data collection on infant rapid weight gain.40 Specifically, parameters on infant’s weight and body length were collected to identify infants with rapid weight gain, and to derive growth curves with innovative Functional Data Analysis techniques.40 Authors revealed that growth curves were negatively associated with the oral microbial diversity, and positively associated with the Firmicutes-to-Bacteroidetes ratio of the oral microbiota.40 The study results suggest for the first time that the association between the oral microbiota and the temporal pattern of weight gain in early childhood might be stronger and more consequential than previously thought, and thus requires further characterization.40 However, the mechanism underlying these associations remains unknown.
Oral microbiome and celiac disease (CD) in children
CD, a chronic immune-mediated enteropathy, affects the small intestinal mucosa after the ingestion of gluten from wheat, rye, and their cross-related varieties in genetically susceptible individuals.145 The estimated prevalence of pediatric CD is between 1/100 and 1/400 (ref. 146). The symptoms of CD in children are characterized by the occurrence of diarrhea, failure to thrive, and abdominal bloating in young infants in the months following gluten introduction.146 Francavilla et al. examined the salivary microbiome and metabolome of 13 CD children with gluten-free diets (T-CD) and their healthy counterparties (HC). Their results suggest that CD is associated with dysbiosis of oral microbiota that could lead to oral metabolome. The salivary level of total cultivable anaerobes significantly (P < 0.05) differed between the T-CD and HC children. Children with T-CD had less diverse salivary microbiome and increased abundance of Rothia, Porphyromonas endodontalis, Gemellaceae, Prevotella nanceiensis, S. sanguinis, and Lachnospiraceae compared to their healthy controls.147 Children with T-CD had a decreased abundance of Actinobacteria, Actinomyces spp., Atopobium spp., and Corynebacterium durum. The relative abundances of these taxa are consistent with higher levels of organic volatile compounds (e.g., ethyl-acetate, nonanal, and 2-hexanone) in the saliva of affected children.147
Oral microbiome and autism in children
Autism spectrum disorder (ASD) reflects a different level of symptom severity in two core domains: deficits in social communication and interaction, and restricted repetitive behaviors.148 ASD occurs in all racial, ethnic, and socioeconomic groups and is more prevalent among boys.148 According to the Center for Disease Control and the Autism and Developmental Disabilities Monitoring Network in the USA, the prevalence is 1 in 88 children.148 With the ASD etiology remaining puzzled, it is particularly important to diagnose and provide appropriate interventions to diseased children at an earlier time point. ASD is associated with several oropharyngeal abnormalities, including buccal sensory sensitivity, taste and texture aversions, speech apraxia, and salivary transcriptome alterations. Alterations in the gut microbiome have been established as features of ASD, which was speculated to impact individual’s behavior via the “microbial–gut–brain axis”.
To uncover the oral microbiome alterations in children with ASD, Hicks et al.149 used shotgun meta-transcriptomic data to identify changes in the salivary microbiome of 348 preschool children who were 2–6 years of age. These children were grouped into three developmental profiles: children with ASD, children with nonautistic developmental delay (DD), and children with typically developing (TD). This research group found five distinguished microbial ratios between ASD and TD children (79.5% accuracy), three distinguished microbial ratios between ASD and DD (76.5% accuracy), and three distinguished microbial ratios between ASD children with and without gastrointestinal disturbance (85.7% accuracy). When comparing the taxa between ASD and TD children, the abundance of two taxa were elevated in ASD children, they were Limnohabitans sp. 63ED37-2 with a false discovery rate (FDR) at 0.01 and Planctomycetales, with a FDR at 0.04. The abundance of four taxa were decreased in ASD children, particulary, Ramlibacter tataouinensis TTB310 with an FDR at 0.001, Mucilaginibacter sp. PAMC 26640 with an FDR at 0.001, Bacteroides vulgatus with an FDR at 0.05, and Gemmata sp. SH-PL17 with an FDR at 0.05. Furthermore, when the taxa abudance between ASD and DD children were compared, two taxa were elevated in children with ASD, Brucella (FDR = 0.05) and Enterococcus faecalis OG1RF (FDR = 0.05), while one taxa (Flavobacterium sp. PK15, FDR = 0.05) was decreased in ASD children. The study results indicate that gut microbiome disruption in ASD could extend to the oropharynx. Future routine assessment of children’s oral microbiome could be developed as a non-invasive and possible sensitive tool to diagnose ASD and assess its progression status.
Oral microbiome and Henoch-Schönlein purpura disease in children
As the most common form of vasculitis in children, Henoch-Schönlein Purpura (HSP) causes inflammation and bleeding in the small blood vessels of the skin, joints, intestines, and kidney.150 Over 75% of children diagnosed with HSP were younger than 10 years old, with a peak incidence at 4–6 years of age.150
In a case–control study, Chen et al.151 examined the association between oral microbiota and HSP by sequencing the 16S rRNA genes of oral samples from 98 HSP children and their 66 healthy counterparts. Distinctive patterns of oral microbiota were seen between the healthy children and those with HSP, higher oral microbial diversity and richness were observed in Children with HSP compared to their controls. Using a linear discriminant analysis (LDA) effect size (LEfSe) algorithm, Chen et al.151 further detected 21 bacterial taxonomic clades showing statistical differences in children with HSP, with 12 increased and 9 decreased taxa. The 12 genera with increased abundance were Neisseriales, Neisseriaceae, Neisseria, Veillonella, Nagativicutes, Veillonellales, Veillonellaceae, Prevotella, Prevotellaceae, Bacteroidetes, Bacteroidia, and Bacteroidales. The nine genera with decreased abundance were Proteobacteria, Gammaproteobacteria, Pseudomonadales, Moraxellaceae, Acinetobacter, Alphaproteobacteria, Pasteurellaceae, Pasteurellales, and Haemophilus.
Notably, the relative abundance of several taxa correlated with clinical manifestations of HSP. The more prolonged hospital stay was associated with a lower abundance of Butyrivibrio sp, but a higher abundance of Haemophilus sp.151 Haemophilus sp was also negatively correlated to the amount of IgE and IgM.151 Prevotella positively correlated with the amount of IgM.151 Prevotella nanceiensis was more abundant in children with HSP and positively correlated with the amount of IgA.151 These study findings that children with HSP have significantly different oral microbiota and a particular association between taxa abundance and HSP clinical parameters exist, suggest the potential of using the oral microbial signatures to identify high-risk populations for HSP and predict clinical progress of HSP children. Although this study does not imply causality between oral microbiota changes and HSP, it provides insight into identifying the types and pathways of bacteria that can be used to predict, prevent, or treat HSP.
Oral microbiome and pediatric appendicitis
Appendicitis is the most common disease that demands urgent surgery among pediatric patients.152 The lifetime risk of developing appendicitis is 7% in girls and 9% in boys.152 Symptoms of appendicitis could range from abdominal pain and vomiting caused by mild or moderate inflammation to life-threatening conditions caused by appendix perforation with extensive contamination.152 Pediatric perforated appendicitis rates are approximately 30% (20%–74%).153 Since younger children have difficulty to articulate their symptoms, a much higher perforation rate is seen among younger children. A cross-sectional study data revealed that the appendix preformation occurred among 100% patients less than 1-year-old and 69% patients at 5 years of age.154
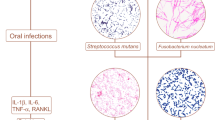
To understand the potential role of oral microbial in pediatric appendicitis, Blod et al.49 rofiled the microbes from inflamed appendices and the gingival sulcus of 21 children who were undergoing appendectomy for acute appendicitis and 28 healthy controls using 16S rRNA sequencing. In addition to profiling the microbial community using sequencing, quantitative measures using RT-qPCR was performed to assess the presence of Fusobacterium nucleatum, Peptostreptococcus stomatis, and Eikenella corrodens. Furthermore, authors used an acid-killing assay to examine these bacteria’s viability to mimic gastric challenge. Although P. stomatis, E. corrodens, and F. nucleatum were detected in both appendicitis and healthy children, more viable P. stomatis were obbserved in the gingival sulci of children with acute appendicitis compared the healthy controls. For children with acute appendicitis, less viable E. corrodens and F. nucleatum presented in inflamed appendices than those found in the gingival sulci. As the oral cavity is the entry port of the gastrointestinal system, the authors proposed a possible oral-gastrointestinal migration of oral bacteria, and subsequently suggest the oral cavity could be a relevant microbial reservoir for developing acute appendicitis.
Oral microbiome and pediatric inflammatory bowel disease
Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the gastrointestinal tract, likely caused by an aberrant immune response to the microbiota and other environmental challenges in genetically susceptible individuals.155 Oral mucosal inflammation is commonly noted in patients with IBD, particularly Crohn’s disease (CD). In all, 0.5%–80% of adult patients with CD manifest oral pathology.156,157 In children, 42% of new diagnoses of CD had oral manifestations.158 Docktor et al.50 examined the oral microbiome (swab samples taken from the tongue and buccal mucosa) from a total of 114 children with CD, ulcerative colitis (UC), and healthy controls, and showed an overall decreased alpha diversity of oral bacterial community in children with CD when compared with healthy controls. In contrast, overall diversity of children with UC did not differ from the healthy controls.
When comparing the tongue samples collected from the CD children to healthy children, several key phyla were significantly reduced, Fusobacteria and Firmicutes.50 When comparing the tongue samples taken from the UC children to the control group, researchers noted a decrease in Fusobacteria in UC children, but an enrichment in Spirochaetes, Synergistetes, and Bacteroidetes.50 No individual phyla from the buccal mucosa samples were significantly different between CD/UC patients and their healthy controls.50 The loss of Fusobacteria and Firmicutes in children with CD were resonated in studies examining the intestinal microbiome.156,158,159 Docktor et al.50 commented that with the prevalence of oral pathology in CD and the ease of oral mucosal sampling, further study could explore the potential of using the oral microbiome as a diagnostic and prognostic tool for pediatric IBD.
Oral microbiome and pediatric obstructive sleep apnea syndrome
As a breathing disorder during sleep, obstructive sleep apnea syndrome (OSAS) is characterized by recurrent episodes of complete or partial upper airway obstruction that interrupt nocturnal ventilation and alter normal sleep patterns.160,161,162 OSAS occurs among children of all age, including infants, with a peak incidence between 3 and 6 years of age.161 The prevalence of pediatric OSAS has been estimated to be approximately 3%.163 While associations between oral diseases and OSAS in the adult population were observed in several cross-sectional studies, limited research reported potential association among the pediatric population.
In a case–control study, Xu et al.164 compared the oral microbiome composition between 30 children OSAS and 30 health counterparts using 16S rRNA gene sequencing. Their results indicate that the oral microbiome composition was significantly different in pediatric OSAS compared to their healthy controls. Thermus, Pseudomonas, Lautropia, and Achromobacter showed higher abundances in the normal control group, whereas Veillonella, Prevotella, Mogibacterium, Campylobacter, and Butyrivibrio were more abundant in the patients with OSAS. In complementary to comparing the oral microbiome, this study group analyzed the urinary metabolome of the study participants and revealed that specific oral microbial changes were correlated with urinary metabolite perturbations in pediatric OSAS. However, the further causal relationship needs examination in longitudinally designed cohort studies. An additional concern lies that there is now ample data confirming that OSAS associated with obesity is highly prevalent in children.165 Whether the imbalanced oral microbiome demonstrated between children with and without OSAS is due to obesity or imbalanced diet that led to obesity remains unknown.
Limitations of current evidence and summary
The mouth is the portal to the gut. Its microbial ecology represents a possible marker if not a risk factor for the disease. The recent advances in salivary biomarker diagnostics and oral microbiome analyses have broadened the discovery of microbial pathogens associated with oral and systematic diseases (e.g., dental caries, periodontal disease, autoimmune diseases, diabetes, and cancers). Although more attention has been paid to the association between gut microbiota and overall health, the oral microbiota has shown its relevance and possibility of being surrogates of gut microbiota, which could provide equivalent dialogistic power with better handling. Compared to the gut microbiome sampling, oral sampling is more psychologically acceptable more easily accessed by patients, as well as by the healthcare professional. Among vulnerable populations, such as infants and young children, oral samples make the perfect diagnostic medium due to its noninvasive and easy handling properties, which holds great promise to be used as diagnostic tools.
With the above-mentioned disease-diagnostic promises, major limitations of establishing the associations between the oral microbiome and children’s diseases lie in:
- (1)
The currently available evidence from the majority of the oral microbiome studies are cross-sectional or case–control designed with a small sample size, which makes it impossible to establish causative associations between oral microbial community and diseases. For example, with the exception of the well-established association established between S. mutans and ECC from various of existing studies across case–control and prospective studies,42,43,44,45,46,47 studies that examined the oral microbiome of children with systemic diseases including pediatric autism, irritable bowel syndrome, pediatric appendicitis, CDs, etc., are lack of power to imply causal relationships between the oral microbiome and the diseases due to the limitation of the study design. Thus, it is not clear whether the change in oral microbial community is a predictor of future disease, or a result of systemic diseases.
- (2)
The current childhood microbiome studies based exclusively on 16S rRNA amplicon sequencing and are therefore limited to the assessment of taxonomic composition and diversity of the microbiome. A detailed metagenomic analysis of microbiota functions that contribute to host-microbiome interactions within the oral environment is needed to identify the mechanistic basis of these interactions and identify metabolic or virulence pathways as therapeutic targets.
- (3)
The majority of oral microbiome studies are focused solely on the bacterial community and do not address critical contributions of the fungal and viral members of the oral microbial community to oral health and disease. Since cross-kingdom interactions have received applause in recent years, given multiple cross-kingdom interactions have been identified (bacteriome and fungi, host and fungi, for say), future research should consider examining the interaction between different microbial community and their interaction with the host.
- (4)
Oral microbiome studies among healthy adults indicate that the diversity of the oral microbiome varies by geography and race/ethnicity166 including inter-social group variation.167 However, few studies have addressed the impact of racial diversity on oral microbiome development in early infancy.
- (5)
Lack of standardized procedure for oral sample collection. To enable microbiota data comparison, standardization of each step in the process for sequencing data generation and analysis is critical. These procedures span from clinical sampling, sample handling and storing, sequencing, bioinformatic data processing, to taxonomic interpretation. One of the most critical factors of the upstream of the process is the oral sample collection. The timing of the sampling (morning, afternoon, before, or after children’s feeding), sample collection tools, sample storing are all critical factors that can induce a significant difference in downstream results. Our collective experience from birth cohort studies that require oral sampling from newborns to toddlers suggests sampling in young children, particularly in toddlers, is even more challenging due to their mobility and limited cooperation. Non-standardized sampling methods could lead to an inherently biased study and challenge the data comparability across study time points internally and across external studies. The development of standardized sample collection methods is urgently needed and would facilitate a qualified comparison of data in the field.
In summary, the complex interplay between the oral microbiome and microbiomes colonized at other body parts in early infancy, host immune factors, and health, suggests complex bidirectional, non-linear interactions that make causality challenging to tease apart making this a very fruitful area of scientific inquiry. The future use of oral microbiome to advance human health will depend on further validation of disease-specific microbial biomarkers and their incorporation into diagnostic and preventive programs that are sensitive and specific, rapid in result delivery and cost-effective for broad implementation. With the complementary of human genomics, proteomics, transcriptomics, metabolomics, the children’s oral microbiome may stand in the center stage of the future precision medicine and personalized medicine.
References
Cho, I. & Blaser, M. J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Machiels, K. et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283 (2014).
De Palma, G. et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 10, 63 (2010).
Tjellstrom, B. et al. Gut microflora associated characteristics in children with celiac disease. Am. J. Gastroenterol. 100, 2784–2788 (2005).
Kelly, J. R. et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 82, 109–118 (2016).
Pistollato, F. et al. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr. Rev. 74, 624–634 (2016).
Schott, E. M. et al. Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight 3, https://doi.org/10.1172/jci.insight.95997 (2018).
Yost, S. et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. Int. J. Oral. Sci. 10, 32 (2018).
Snider, E. J. et al. Barrett’s esophagus is associated with a distinct oral microbiome. Clin. Transl. Gastroenterol. 9, 135 (2018).
Shillitoe, E. J. The microbiome of oral cancer. Crit. Rev. Oncog. 23, 153–160 (2018).
Klimesova, K., Jiraskova Zakostelska, Z. & Tlaskalova-Hogenova, H. Oral bacterial and fungal microbiome impacts colorectal carcinogenesis. Front. Microbiol. 9, 774 (2018).
Gaiser, R. A. et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 68, 2186–2194 (2019).
Brennan, C. A. & Garrett, W. S. Fusobacterium nucleatum—symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166 (2019).
Lockhart, P. B. et al. Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association?: a scientific statement from the American Heart Association. Circulation 125, 2520–2544 (2012).
van de Wijgert, J. & Jespers, V. The global health impact of vaginal dysbiosis. Res. Microbiol. 168, 859–864 (2017).
Borgdorff, H. et al. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal Immunol. 9, 621–633 (2016).
Lewis, F. M., Bernstein, K. T. & Aral, S. O. Vaginal microbiome and its relationship to behavior, sexual health, and sexually transmitted diseases. Obstet. Gynecol. 129, 643–654 (2017).
van de Wijgert, J. H. et al. The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS ONE 9, e105998 (2014).
van de Wijgert, J. The vaginal microbiome and sexually transmitted infections are interlinked: consequences for treatment and prevention. PLoS Med. 14, e1002478 (2017).
Backhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 852 (2015).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Grier, A. et al. Neonatal gut and respiratory microbiota: coordinated development through time and space. Microbiome 6, 193 (2018).
Grier, A. et al. Impact of prematurity and nutrition on the developing gut microbiome and preterm infant growth. Microbiome 5, 158 (2017).
Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Koenig, J. E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108, 4578–4585 (2011).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Mason, M. R., Chambers, S., Dabdoub, S. M., Thikkurissy, S. & Kumar, P. S. Characterizing oral microbial communities across dentition states and colonization niches. Microbiome 6, 67 (2018).
Nelson-Filho, P. et al. Dynamics of microbial colonization of the oral cavity in newborns. Braz. Dent. J. 24, 415–419 (2013).
Rotimi, V. O. & Duerden, B. I. The development of the bacterial flora in normal neonates. J. Med. Microbiol. 14, 51–62 (1981).
Ward, T. L. et al. Development of the human mycobiome over the first month of life and across body sites. mSystems 3, https://doi.org/10.1128/mSystems.00140-17 (2018).
Escapa, I. F., Chen, T., Yanmei Huang, Y., Gajare, P., Dewhirst F. E. & Lemon, K. P. New insights into human nostril microbiome from the expanded Human Oral Microbiome Database (eHOMD): a resource for species-level identification of microbiome data from the aerodigestive tract. mSystems 3, 00187–18 (2018).
Yu, J. C. et al. Innate immunity of neonates and infants. Front. Immunol. 9, 1759 (2018).
Wu, R. Q., Zhang, D. F., Tu, E., Chen, Q. M. & Chen, W. The mucosal immune system in the oral cavity-an orchestra of T cell diversity. Int. J. Oral. Sci. 6, 125–132 (2014).
Backhed, F. Host responses to the human microbiome. Nutr. Rev. 70(Suppl 1), S14–S17 (2012).
La Rosa, P. S. et al. Patterned progression of bacterial populations in the premature infant gut. Proc. Natl Acad. Sci. USA 111, 12522–12527 (2014).
White, R. A. et al. Novel developmental analyses identify longitudinal patterns of early gut microbiota that affect infant growth. PLoS Comput. Biol. 9, e1003042 (2013).
Derrien, M. et al. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1, 254–268 (2010).
Craig, S. J. C. et al. Child weight gain trajectories linked to oral microbiota composition. Sci. Rep. 8, 14030 (2018).
Malamud, D. Saliva as a diagnostic fluid. Dent. Clin. North Am. 55, 159–178 (2011).
Gross, E. L. et al. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS ONE 7, e47722 (2012).
Johansson, I., Witkowska, E., Kaveh, B., Lif Holgerson, P. & Tanner, A. C. The microbiome in populations with a low and high prevalence of caries. J. Dent. Res. 95, 80–86 (2016).
Ma, C. et al. Comparison of oral microbial profiles between children with severe early childhood caries and caries-free children using the human oral microbe identification microarray. PLoS ONE 10, e0122075 (2015).
Richards, V. P. et al. Microbiomes of site-specific dental plaques from children with different caries status. Infect. Immun. 85, https://doi.org/10.1128/IAI.00106-17 (2017).
Tanner, A. C. et al. Microbiota of severe early childhood caries before and after therapy. J. Dent. Res. 90, 1298–1305 (2011).
Yang, F. et al. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J. 6, 1–10 (2012).
Xiao, J. et al. Association between oral candida and bacteriome in children with severe ECC. J. Dent. Res. 97, 1468–1476 (2018).
Blod, C. et al. The oral microbiome-the relevant reservoir for acute pediatric appendicitis? Int. J. Colorectal Dis. 33, 209–218 (2018).
Docktor, M. J. et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 18, 935–942 (2012).
Harkins, C. P., Kong, H. H. & Segre, J. A. Manipulating the human microbiome to manage disease. JAMA https://doi.org/10.1001/jama.2019.19602 (2019).
Panigrahi, P. et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412 (2017).
Aagaard, K. et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra265 (2014).
Bearfield, C., Davenport, E. S., Sivapathasundaram, V. & Allaker, R. P. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG 109, 527–533 (2002).
Gomez-Arango, L. F. et al. Contributions of the maternal oral and gut microbiome to placental microbial colonization in overweight and obese pregnant women. Sci. Rep. 7, 2860 (2017).
Mold, J. E. et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science 322, 1562–1565 (2008).
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Gronlund, M. M., Lehtonen, O. P., Eerola, E. & Kero, P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J. Pediatr. Gastroenterol. Nutr. 28, 19–25 (1999).
Dzidic, M. et al. Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 12, 2292–2306 (2018).
Boix-Amoros, A., Collado, M. C. & Mira, A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 7, 492 (2016).
Merglova, V. & Polenik, P. Early colonization of the oral cavity in 6- and 12-month-old infants by cariogenic and periodontal pathogens: a case-control study. Folia Microbiol. (Praha) 61, 423–429 (2016).
Hegde, S. & Munshi, A. K. Influence of the maternal vaginal microbiota on the oral microbiota of the newborn. J. Clin. Pediatr. Dent. 22, 317–321 (1998).
Gomez, A. & Nelson, K. E. The oral microbiome of children: development, disease, and implications beyond oral health. Microb. Ecol. 73, 492–503 (2017).
Cephas, K. D. et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS ONE 6, e23503 (2011).
Crielaard, W. et al. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med. Genomics 4, 22 (2011).
Kleinegger, C. L., Lockhart, S. R., Vargas, K. & Soll, D. R. Frequency, intensity, species, and strains of oral Candida vary as a function of host age. J. Clin. Microbiol. 34, 2246–2254 (1996).
Darwazeh, A. M. & al-Bashir, A. Oral candidal flora in healthy infants. J. Oral. Pathol. Med. 24, 361–364 (1995).
Stecksen-Blicks, C., Granstrom, E., Silfverdal, S. A. & West, C. E. Prevalence of oral Candida in the first year of life. Mycoses 58, 550–556 (2015).
Scully, C., el-Kabir, M. & Samaranayake, L. P. Candida and oral candidosis: a review. Crit. Rev. Oral. Biol. Med. 5, 125–157 (1994).
Ghannoum, M. A. et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6, e1000713 (2010).
Pride, D. T. et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 6, 915–926 (2012).
Corstjens, P. L., Abrams, W. R. & Malamud, D. Saliva and viral infections. Periodontology 2000 70, 93–110, https://doi.ort/10.1111/prd.12112 (2016).
Parras-Molto, M., Suarez-Rodriguez, P., Eguia, A., Aguirre-Urizar, J. M. & Lopez-Bueno, A. Genome sequence of two novel species of torque teno minivirus from the human oral cavity. Genome Announc. 2, https://doi.org/10.1128/genomeA.00868-14 (2014).
Pinninti, S. G. & Kimberlin, D. W. Neonatal herpes simplex virus infections. Semin. Perinatol. 42, 168–175 (2018).
Mahnert, N., Roberts, S. W., Laibl, V. R., Sheffield, J. S. & Wendel, G. D. Jr The incidence of neonatal herpes infection. Am. J. Obstet. Gynecol. 196, e55–e56 (2007).
Kimberlin, D. W. et al. Natural history of neonatal herpes simplex virus infections in the acyclovir era. Pediatrics 108, 223–229 (2001).
Sallberg, M. Oral viral infections of children. Periodontol 2000 49, 87–95 (2009).
Gao, L. et al. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell, https://doi.org/10.1007/s13238-018-0548-1 (2018).
Ward, T. L., Knights, D. & Gale, C. A. Infant fungal communities: current knowledge and research opportunities. BMC Med. 15, 30 (2017).
Chen, P. E. & Shapiro, B. J. The advent of genome-wide association studies for bacteria. Curr. Opin. Microbiol. 25, 17–24 (2015).
Demmitt, B. A. et al. Genetic influences on the human oral microbiome. BMC Genomics 18, 659 (2017).
Gomez, A. et al. Host genetic control of the oral microbiome in health and disease. Cell Host Microbe 22, 269–278 e263 (2017).
Blekhman, R. et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16, 191 (2015).
Bretz, W. A. et al. Dental caries and microbial acid production in twins. Caries Res. 39, 168–172 (2005).
Abusleme, L. et al. Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI Insight 3, https://doi.org/10.1172/jci.insight.122061 (2018).
Holland, S. M. et al. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357, 1608–1619 (2007).
Minegishi, Y. et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448, 1058–1062 (2007).
World Health Organization. Preterm Birth (2018).
Liu, L. et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the sustainable development goals. Lancet 388, 3027–3035 (2016).
Younge, N. E., Araujo-Perez, F., Brandon, D. & Seed, P. C. Early-life skin microbiota in hospitalized preterm and full-term infants. Microbiome 6, 98 (2018).
Sohn, K., Kalanetra, K. M., Mills, D. A. & Underwood, M. A. Buccal administration of human colostrum: impact on the oral microbiota of premature infants. J. Perinatol. 36, 106–111 (2016).
Underwood, M. A. & Sohn, K. The microbiota of the extremely preterm infant. Clin. Perinatol. 44, 407–427 (2017).
Hewitt, K. M. et al. Bacterial diversity in two Neonatal Intensive Care Units (NICUs). PLoS ONE 8, e54703 (2013).
Bokulich, N. A., Mills, D. A. & Underwood, M. A. Surface microbes in the neonatal intensive care unit: changes with routine cleaning and over time. J. Clin. Microbiol. 51, 2617–2624 (2013).
Hartz, L. E., Bradshaw, W. & Brandon, D. H. Potential NICU environmental influences on the neonate’s microbiome: a systematic review. Adv. Neonatal Care 15, 324–335 (2015).
Ferraris, L. et al. Clostridia in premature neonates’ gut: incidence, antibiotic susceptibility, and perinatal determinants influencing colonization. PLoS ONE 7, e30594 (2012).
Gomez-Arango, L. F. et al. Antibiotic treatment at delivery shapes the initial oral microbiome in neonates. Sci. Rep. 7, 43481 (2017).
Lif Holgerson, P., Harnevik, L., Hernell, O., Tanner, A. C. & Johansson, I. Mode of birth delivery affects oral microbiota in infants. J. Dent. Res. 90, 1183–1188 (2011).
Drell, T. et al. The influence of different maternal microbial communities on the development of infant gut and oral microbiota. Sci. Rep. 7, 9940 (2017).
Hurley, E. et al. The microbiota of the mother at birth and its influence on the emerging infant oral microbiota from birth to 1 year of age: a cohort study. J. Oral. Microbiol. 11, 1599652 (2019).
Kennedy, B. et al. Oral microbiota development in early childhood. Sci. Rep. 9, 19025 (2019).
Nuriel-Ohayon, M., Neuman, H. & Koren, O. Microbial changes during pregnancy, birth, and infancy. Front. Microbiol. 7, 1031 (2016).
Lee, S. A. et al. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr. Res. Pract. 9, 242–248 (2015).
Holgerson, P. L. et al. Oral microbial profile discriminates breast-fed from formula-fed infants. J. Pediatr. Gastroenterol. Nutr. 56, 127–136 (2013).
Goldsmith, F., O’Sullivan, A., Smilowitz, J. T. & Freeman, S. L. Lactation and intestinal microbiota: how early diet shapes the infant gut. J. Mammary Gland Biol. Neoplasia 20, 149–158 (2015).
Danielsson Niemi, L., Hernell, O. & Johansson, I. Human milk compounds inhibiting adhesion of mutans Streptococci to host ligand-coated hydroxyapatite in vitro. Caries Res. 43, 171–178 (2009).
Sweeney, E. L. et al. The effect of breastmilk and saliva combinations on the in vitro growth of oral pathogenic and commensal microorganisms. Sci. Rep. 8, 15112 (2018).
Timby, N. et al. Oral microbiota in infants fed a formula supplemented with bovine milk fat globule membranes—a randomized controlled trial. PLoS ONE 12, e0169831 (2017).
Parisotto, T. M., Steiner-Oliveira, C., Silva, C. M., Rodrigues, L. K. & Nobre-dos-Santos, M. Early childhood caries and mutans streptococci: a systematic review. Oral. Health Prev. Dent. 8, 59–70 (2010).
Wernersson, J., Danielsson Niemi, L., Einarson, S., Hernell, O. & Johansson, I. Effects of human milk on adhesion of Streptococcus mutans to saliva-coated hydroxyapatite in vitro. Caries Res. 40, 412–417 (2006).
Avila, W. M., Pordeus, I. A., Paiva, S. M. & Martins, C. C. Breast and bottle feeding as risk factors for dental caries: a systematic review and meta-analysis. PLoS ONE 10, e0142922 (2015).
Plonka, K. A. et al. A longitudinal study comparing mutans streptococci and lactobacilli colonisation in dentate children aged 6 to 24 months. Caries Res. 46, 385–393 (2012).
Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145 e135 (2018).
Filippidi, A. et al. The effect of maternal flora on Candida colonisation in the neonate. Mycoses 57, 43–48 (2014).
Al-Rusan, R. M., Darwazeh, A. M. & Lataifeh, I. M. The relationship of Candida colonization of the oral and vaginal mucosae of mothers and oral mucosae of their newborns at birth. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 123, 459–463 (2017).
Teughels, W. et al. Bacteria interfere with A. actinomycetemcomitans colonization. J. Dent. Res. 86, 611–617 (2007).
Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 (2017).
Bliss, J. M., Basavegowda, K. P., Watson, W. J., Sheikh, A. U. & Ryan, R. M. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr. Infect. Dis. J. 27, 231–235 (2008).
Waggoner-Fountain, L. A. et al. Vertical and horizontal transmission of unique Candida species to premature newborns. Clin. Infect. Dis. 22, 803–808 (1996).
Xiao, J. et al. Candida albicans carriage in children with severe early childhood caries (S-ECC) and maternal relatedness. PLoS ONE 11, e0164242 (2016).
Yang, Y. et al. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int. J. Cancer, https://doi.org/10.1002/ijc.31941 (2018).
Sun, J. H. et al. A screening method for gastric cancer by oral microbiome detection. Oncol. Rep. 39, 2217–2224 (2018).
Peters, B. A. et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 77, 6777–6787 (2017).
Bracci, P. M. Oral health and the oral microbiome in pancreatic cancer: an overview of epidemiological studies. Cancer J. 23, 310–314 (2017).
Galvao-Moreira, L. V. & da Cruz, M. C. Oral microbiome, periodontitis and risk of head and neck cancer. Oral. Oncol. 53, 17–19 (2016).
Fourie, N. H. et al. The microbiome of the oral mucosa in irritable bowel syndrome. Gut Microbes 7, 286–301 (2016).
He, J., Li, Y., Cao, Y., Xue, J. & Zhou, X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol. (Praha) 60, 69–80 (2015).
Vos, T. A. A. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259 (2017).
Dye, B. A., Li, X. & Thorton-Evans, G. Oral health disparities as determined by selected healthy people 2020 oral health objectives for the United States, 2009-2010. NCHS Data Brief, 1–8 (2012).
Kanasi, E. et al. Microbial risk markers for childhood caries in pediatricians’ offices. J. Dent. Res. 89, 378–383 (2010).
Caufield, P. W., Cutter, G. R. & Dasanayake, A. P. Initial acquisition of mutans streptococci by infants: evidence for a discrete window of infectivity. J. Dent. Res. 72, 37–45 (1993).
Li, Y., Caufield, P. W., Dasanayake, A. P., Wiener, H. W. & Vermund, S. H. Mode of delivery and other maternal factors influence the acquisition of Streptococcus mutans in infants. J. Dent. Res. 84, 806–811 (2005).
Zhan, L., Tan, S., Den Besten, P., Featherstone, J. D. & Hoover, C. I. Factors related to maternal transmission of mutans streptococci in high-risk children-pilot study. Pediatr. Dent. 34, e86–e91 (2012).
Slayton, R. L. Reducing mutans streptococci levels in caregivers may reduce transmission to their children and lead to reduced caries prevalence. J. Evid. Based Dent. Pract. 11, 27–28 (2011).
Klein, M. I., Florio, F. M., Pereira, A. C., Hofling, J. F. & Goncalves, R. B. Longitudinal study of transmission, diversity, and stability of Streptococcus mutans and Streptococcus sobrinus genotypes in Brazilian nursery children. J. Clin. Microbiol. 42, 4620–4626 (2004).
Klinke, T. et al. Changes in Candida spp., mutans streptococci and lactobacilli following treatment of early childhood caries: a 1-year follow-up. Caries Res. 48, 24–31 (2014).
Plonka, K. A. et al. Mutans streptococci and lactobacilli colonization in predentate children from the neonatal period to seven months of age. Caries Res. 46, 213–220 (2012).
Adler, C. J. et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 45, 455e451 (2013).
Fontana, M. et al. Identification of caries risk factors in toddlers. J. Dent. Res. 90, 209–214 (2011).
Piva, F. et al. A longitudinal study of early childhood caries and associated factors in Brazilian. Child. Braz. Dent. J. 28, 241–248 (2017).
Xiao, J. et al. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 52, 102–112 (2018).
Bowen, W. H. & Koo, H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86 (2011).
Caufield, P. W. et al. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68, 4018–4023 (2000).
Tye-Din, J. & Anderson, R. Immunopathogenesis of celiac disease. Curr. Gastroenterol. Rep. 10, 458–465 (2008).
Garnier-Lengline, H., Cerf-Bensussan, N. & Ruemmele, F. M. Celiac disease in children. Clin. Res. Hepatol. Gastroenterol. 39, 544–551 (2015).
Francavilla, R. et al. Salivary microbiota and metabolome associated with celiac disease. Appl. Environ. Microbiol. 80, 3416–3425 (2014).
Ivanov, H. Y., Stoyanova, V. K., Popov, N. T. & Vachev, T. I. Autism spectrum disorder—a complex genetic disorder. Folia Med. (Plovdiv.) 57, 19–28 (2015).
Hicks, S. D. et al. Oral microbiome activity in children with autism spectrum disorder. Autism Res. 11, 1286–1299 (2018).
Pavlovic, M., Radlovic, N., Lekovic, Z., Berenji, K. & Novak, A. [Corticosteroid therapy in Henoch-Schonlein gastritis]. Srp. Arh. Celok. Lek. 135, 208–211 (2007).
Chen, B. et al. Oral microbiota dysbiosis and its association with Henoch-Schonlein Purpura in children. Int. Immunopharmacol. 65, 295–302 (2018).
Guthery, S. L., Hutchings, C., Dean, J. M. & Hoff, C. National estimates of hospital utilization by children with gastrointestinal disorders: analysis of the 1997 kids’ inpatient database. J. Pediatr. 144, 589–594 (2004).
Addiss, D. G., Shaffer, N., Fowler, B. S. & Tauxe, R. V. The epidemiology of appendicitis and appendectomy in the United States. Am. J. Epidemiol. 132, 910–925 (1990).
Nance, M. L., Adamson, W. T. & Hedrick, H. L. Appendicitis in the young child: a continuing diagnostic challenge. Pediatr. Emerg. Care 16, 160–162 (2000).
Kaser, A., Zeissig, S. & Blumberg, R. S. Inflammatory bowel disease. Annu. Rev. Immunol. 28, 573–621 (2010).
Rowland, M., Fleming, P. & Bourke, B. Looking in the mouth for Crohn’s disease. Inflamm. Bowel Dis. 16, 332–337 (2010).
Galbraith, S. S., Drolet, B. A., Kugathasan, S., Paller, A. S. & Esterly, N. B. Asymptomatic inflammatory bowel disease presenting with mucocutaneous findings. Pediatrics 116, e439–e444 (2005).
Harty, S. et al. A prospective study of the oral manifestations of Crohn’s disease. Clin. Gastroenterol. Hepatol. 3, 886–891 (2005).
Pittock, S. et al. The oral cavity in Crohn’s disease. J. Pediatr. 138, 767–771 (2001).
Standards and indications for cardiopulmonary sleep studies in children. American Thoracic Society. Am. J. Respir. Crit. Care Med. 153, 866–878 (1996).
Marcus, C. L. & Loughlin, G. M. Obstructive sleep apnea in children. Semin. Pediatr. Neurol. 3, 23–28 (1996).
Ierardo, G., Luzzi, V. & Polimeni, A. Obstructive Sleep Apnea Syndrome (OSAS): evaluation and treatment of odontostomatological problems. Med Lav. 108, 293–296 (2017).
Chang, S. J. & Chae, K. Y. Obstructive sleep apnea syndrome in children: epidemiology, pathophysiology, diagnosis and sequelae. Korean J. Pediatr. 53, 863–871 (2010).
Xu, H. et al. Pediatric obstructive sleep apnea is associated with changes in the oral microbiome and urinary metabolomics profile: a pilot study. J. Clin. Sleep. Med. 14, 1559–1567 (2018).
Narang, I. & Mathew, J. L. Childhood obesity and obstructive sleep apnea. J. Nutr. Metab. 2012, 134202 (2012).
Gupta, V. K., Paul, S. & Dutta, C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 8, 1162 (2017).
Song, S. J. et al. Cohabiting family members share microbiota with one another and with their dogs. Elife 2, e00458 (2013).
Acknowledgements
J.X.’s study is supported by the National Institute for Dental and Craniofacial Research K23 DE027412. The funding agencies had no role in the study design, data collection, analyses, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.X., K.F., and S.R.G. contributed to the manuscript conception, design, drafting, and critical revision. All authors gave final approval and agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xiao, J., Fiscella, K.A. & Gill, S.R. Oral microbiome: possible harbinger for children’s health. Int J Oral Sci 12, 12 (2020). https://doi.org/10.1038/s41368-020-0082-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41368-020-0082-x
This article is cited by
-
The tongue microbiome of young patients with chronic kidney disease and their healthy mothers
Clinical Oral Investigations (2024)
-
Metagenomic analysis examines oral microbiome changes and interplay with immune response following prenatal total oral rehabilitation
Journal of Translational Medicine (2023)
-
The microbiota of pregnant women with SARS-CoV-2 and their infants
Microbiome (2023)
-
The breast milk and childhood gastrointestinal microbiotas and disease outcomes: a longitudinal study
Pediatric Research (2023)
-
The association of tonsillar microbiota with biochemical indices based on obesity and tonsillar hypertrophy in children
Scientific Reports (2023)